Single-cell analysis of Daxx and ATRX-dependent transcriptional repression
- PMID: 22976303
- PMCID: PMC3561858
- DOI: 10.1242/jcs.110148
Single-cell analysis of Daxx and ATRX-dependent transcriptional repression
Abstract
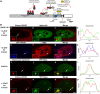
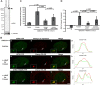
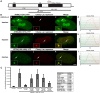
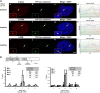
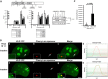
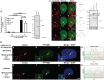
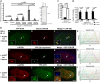
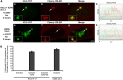
Histone H3.3 is a constitutively expressed H3 variant implicated in the epigenetic inheritance of chromatin structures. Recently, the PML-nuclear body (PML-NB)/Nuclear Domain 10 (ND10) proteins, Daxx and ATRX, were found to regulate replication-independent histone H3.3 chromatin assembly at telomeres and pericentric heterochromatin. As it is not completely understood how PML-NBs/ND10s regulate transcription and resistance to viral infection, we have used a CMV-promoter-regulated inducible transgene array, at which Daxx and ATRX are enriched, to delineate the mechanisms through which they regulate transcription. When integrated into HeLa cells, which express both Daxx and ATRX, the array is refractory to activation. However, transcription can be induced when ICP0, the HSV-1 E3 ubiquitin ligase required to reverse latency, is expressed. As ATRX and Daxx are depleted from the activated array in ICP0-expressing HeLa cells, this suggests that they are required to maintain a repressed chromatin environment. As histone H3.3 is strongly recruited to the ICP0-activated array but does not co-localize with the DNA, this also suggests that chromatin assembly is blocked during activation. The conclusion that the Daxx and ATRX pathway is required for transcriptional repression and chromatin assembly at this site is further supported by the finding that an array integrated into the ATRX-negative U2OS cell line can be robustly activated and that histone H3.3 is similarly recruited and unincorporated into the chromatin. Therefore, this study has important implications for understanding gene silencing, viral latency and PML-NB/ND10 function.
Figures
Similar articles
-
Sp100A promotes chromatin decondensation at a cytomegalovirus-promoter-regulated transcription site.Mol Biol Cell. 2013 May;24(9):1454-68. doi: 10.1091/mbc.E12-09-0669. Epub 2013 Mar 13. Mol Biol Cell. 2013. PMID: 23485562 Free PMC article.
-
Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes.Nucleic Acids Res. 2013 Apr 1;41(6):3532-50. doi: 10.1093/nar/gkt064. Epub 2013 Feb 8. Nucleic Acids Res. 2013. PMID: 23396441 Free PMC article.
-
PML is recruited to heterochromatin during S phase and represses DAXX-mediated histone H3.3 chromatin assembly.J Cell Sci. 2019 Mar 26;132(6):jcs220970. doi: 10.1242/jcs.220970. J Cell Sci. 2019. PMID: 30796101 Free PMC article.
-
New players in heterochromatin silencing: histone variant H3.3 and the ATRX/DAXX chaperone.Nucleic Acids Res. 2016 Feb 29;44(4):1496-501. doi: 10.1093/nar/gkw012. Epub 2016 Jan 14. Nucleic Acids Res. 2016. PMID: 26773061 Free PMC article. Review.
-
Maintaining memory of silencing at imprinted differentially methylated regions.Cell Mol Life Sci. 2016 May;73(9):1871-9. doi: 10.1007/s00018-016-2157-6. Epub 2016 Feb 16. Cell Mol Life Sci. 2016. PMID: 26883803 Free PMC article. Review.
Cited by
-
Multiple Roles of dXNP and dADD1-Drosophila Orthologs of ATRX Chromatin Remodeler.Int J Mol Sci. 2023 Nov 18;24(22):16486. doi: 10.3390/ijms242216486. Int J Mol Sci. 2023. PMID: 38003676 Free PMC article. Review.
-
HSV-1 ICP0: An E3 Ubiquitin Ligase That Counteracts Host Intrinsic and Innate Immunity.Cells. 2014 May 20;3(2):438-54. doi: 10.3390/cells3020438. Cells. 2014. PMID: 24852129 Free PMC article.
-
Single cell analysis of RNA-mediated histone H3.3 recruitment to a cytomegalovirus promoter-regulated transcription site.J Biol Chem. 2013 Jul 5;288(27):19882-99. doi: 10.1074/jbc.M113.473181. Epub 2013 May 20. J Biol Chem. 2013. PMID: 23689370 Free PMC article.
-
Sp100A promotes chromatin decondensation at a cytomegalovirus-promoter-regulated transcription site.Mol Biol Cell. 2013 May;24(9):1454-68. doi: 10.1091/mbc.E12-09-0669. Epub 2013 Mar 13. Mol Biol Cell. 2013. PMID: 23485562 Free PMC article.
-
Multifunctional adaptor protein Daxx interacts with chromatin-remodelling ATPase Brg1.Biochem Biophys Rep. 2015 Dec 30;5:246-252. doi: 10.1016/j.bbrep.2015.12.012. eCollection 2016 Mar. Biochem Biophys Rep. 2015. PMID: 28955830 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

