Differential clade-specific HLA-B*3501 association with HIV-1 disease outcome is linked to immunogenicity of a single Gag epitope
- PMID: 22973023
- PMCID: PMC3497693
- DOI: 10.1128/JVI.01381-12
Differential clade-specific HLA-B*3501 association with HIV-1 disease outcome is linked to immunogenicity of a single Gag epitope
Abstract
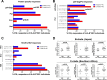
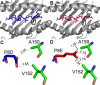
The strongest genetic influence on immune control in HIV-1 infection is the HLA class I genotype. Rapid disease progression in B-clade infection has been linked to HLA-B*35 expression, in particular to the less common HLA-B*3502 and HLA-B*3503 subtypes but also to the most prevalent subtype, HLA-B*3501. In these studies we first demonstrated that whereas HLA-B*3501 is associated with a high viral set point in two further B-clade-infected cohorts, in Japan and Mexico, this association does not hold in two large C-clade-infected African cohorts. We tested the hypothesis that clade-specific differences in HLA associations with disease outcomes may be related to distinct targeting of critical CD8(+) T-cell epitopes. We observed that only one epitope was significantly targeted differentially, namely, the Gag-specific epitope NPPIPVGDIY (NY10, Gag positions 253 to 262) (P = 2 × 10(-5)). In common with two other HLA-B*3501-restricted epitopes, in Gag and Nef, that were not targeted differentially, a response toward NY10 was associated with a significantly lower viral set point. Nonimmunogenicity of NY10 in B-clade-infected subjects derives from the Gag-D260E polymorphism present in ∼90% of B-clade sequences, which critically reduces recognition of the Gag NY10 epitope. These data suggest that in spite of any inherent HLA-linked T-cell receptor repertoire differences that may exist, maximizing the breadth of the Gag-specific CD8(+) T-cell response, by the addition of even a single epitope, may be of overriding importance in achieving immune control of HIV infection. This distinction is of direct relevance to development of vaccines designed to optimize the anti-HIV CD8(+) T-cell response in all individuals, irrespective of HLA type.
Figures





Similar articles
-
HIV subtype influences HLA-B*07:02-associated HIV disease outcome.AIDS Res Hum Retroviruses. 2014 May;30(5):468-75. doi: 10.1089/AID.2013.0197. Epub 2013 Oct 4. AIDS Res Hum Retroviruses. 2014. PMID: 24010680 Free PMC article.
-
Immunological control of chronic HIV-1 infection: HLA-mediated immune function and viral evolution in adolescents.AIDS. 2007 Nov 30;21(18):2387-97. doi: 10.1097/QAD.0b013e3282f13823. AIDS. 2007. PMID: 18025875 Free PMC article.
-
HLA-B*57 Micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control.J Virol. 2012 Jan;86(2):919-29. doi: 10.1128/JVI.06150-11. Epub 2011 Nov 16. J Virol. 2012. PMID: 22090105 Free PMC article.
-
The influence of HLA/HIV genetics on the occurrence of elite controllers and a need for therapeutics geotargeting view.Braz J Infect Dis. 2021 Sep-Oct;25(5):101619. doi: 10.1016/j.bjid.2021.101619. Epub 2021 Sep 22. Braz J Infect Dis. 2021. PMID: 34562387 Free PMC article. Review.
-
Protective HLA-B57: T cell and natural killer cell recognition in HIV infection.Biochem Soc Trans. 2022 Oct 31;50(5):1329-1339. doi: 10.1042/BST20220244. Biochem Soc Trans. 2022. PMID: 36111814 Free PMC article. Review.
Cited by
-
Understanding the mechanisms driving the spread of subtype C HIV-1.EBioMedicine. 2020 Mar;53:102682. doi: 10.1016/j.ebiom.2020.102682. Epub 2020 Feb 27. EBioMedicine. 2020. PMID: 32114391 Free PMC article. Review.
-
Molecular insights into the HLA-B35 molecules' classification associated with HIV control.Immunol Cell Biol. 2024 Jan;102(1):34-45. doi: 10.1111/imcb.12698. Epub 2023 Oct 9. Immunol Cell Biol. 2024. PMID: 37811811 Free PMC article.
-
Role of HLA Adaptation in HIV Evolution.Front Immunol. 2016 Jan 18;6:665. doi: 10.3389/fimmu.2015.00665. eCollection 2015. Front Immunol. 2016. PMID: 26834742 Free PMC article. Review.
-
Epitope length variants balance protective immune responses and viral escape in HIV-1 infection.Cell Rep. 2022 Mar 1;38(9):110449. doi: 10.1016/j.celrep.2022.110449. Cell Rep. 2022. PMID: 35235807 Free PMC article.
-
Third-party cytomegalovirus-specific T cells improved survival in refractory cytomegalovirus viremia after hematopoietic transplant.J Clin Invest. 2023 May 15;133(10):e165476. doi: 10.1172/JCI165476. J Clin Invest. 2023. PMID: 36951958 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

