Identifying viral parameters from in vitro cell cultures
- PMID: 22969758
- PMCID: PMC3432869
- DOI: 10.3389/fmicb.2012.00319
Identifying viral parameters from in vitro cell cultures
Abstract
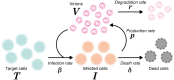
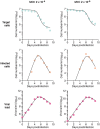
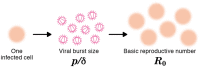
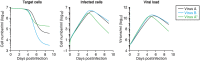
Current in vitro cell culture studies of viral replication deliver detailed time courses of several virological variables, like the amount of virions and the number of target cells, measured over several days of the experiment. Each of these time points solely provides a snap-shot of the virus infection kinetics and is brought about by the complex interplay of target cell infection, and viral production and cell death. It remains a challenge to interpret these data quantitatively and to reveal the kinetics of these underlying processes to understand how the viral infection depends on these kinetic properties. In order to decompose the kinetics of virus infection, we introduce a method to "quantitatively" describe the virus infection in in vitro cell cultures, and discuss the potential of the mathematical based analyses for experimental virology.
Keywords: in vitro experiment; mathematical modeling; quantification; virus infection.
Figures
Similar articles
-
Quantification system for the viral dynamics of a highly pathogenic simian/human immunodeficiency virus based on an in vitro experiment and a mathematical model.Retrovirology. 2012 Feb 25;9:18. doi: 10.1186/1742-4690-9-18. Retrovirology. 2012. PMID: 22364292 Free PMC article.
-
Molecular biological assessment methods and understanding the course of the HIV infection.APMIS Suppl. 2003;(114):1-37. APMIS Suppl. 2003. PMID: 14626050 Review.
-
Modelling Degradation and Replication Kinetics of the Zika Virus In Vitro Infection.Viruses. 2020 May 15;12(5):547. doi: 10.3390/v12050547. Viruses. 2020. PMID: 32429277 Free PMC article.
-
Production of Defective Interfering Particles of Influenza A Virus in Parallel Continuous Cultures at Two Residence Times-Insights From qPCR Measurements and Viral Dynamics Modeling.Front Bioeng Biotechnol. 2019 Oct 18;7:275. doi: 10.3389/fbioe.2019.00275. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31681751 Free PMC article.
-
Mathematical Analysis of Viral Replication Dynamics and Antiviral Treatment Strategies: From Basic Models to Age-Based Multi-Scale Modeling.Front Microbiol. 2018 Jul 11;9:1546. doi: 10.3389/fmicb.2018.01546. eCollection 2018. Front Microbiol. 2018. PMID: 30050523 Free PMC article. Review.
Cited by
-
Quantification of deaminase activity-dependent and -independent restriction of HIV-1 replication mediated by APOBEC3F and APOBEC3G through experimental-mathematical investigation.J Virol. 2014 May;88(10):5881-7. doi: 10.1128/JVI.00062-14. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623435 Free PMC article.
-
Competitive exclusion during co-infection as a strategy to prevent the spread of a virus: A computational perspective.PLoS One. 2021 Feb 24;16(2):e0247200. doi: 10.1371/journal.pone.0247200. eCollection 2021. PLoS One. 2021. PMID: 33626106 Free PMC article.
-
Genomics and computational science for virus research.Front Microbiol. 2013 Mar 7;4:42. doi: 10.3389/fmicb.2013.00042. eCollection 2013. Front Microbiol. 2013. PMID: 23472060 Free PMC article. No abstract available.
-
A highly pathogenic simian/human immunodeficiency virus effectively produces infectious virions compared with a less pathogenic virus in cell culture.Theor Biol Med Model. 2017 Apr 21;14(1):9. doi: 10.1186/s12976-017-0055-8. Theor Biol Med Model. 2017. PMID: 28431573 Free PMC article.
-
Quantification of the dynamics of enterovirus 71 infection by experimental-mathematical investigation.J Virol. 2013 Jan;87(1):701-5. doi: 10.1128/JVI.01453-12. Epub 2012 Oct 24. J Virol. 2013. PMID: 23097444 Free PMC article.
References
-
- Anderson R. M. (1991). The Kermack-McKendrick epidemic threshold theorem. Bull. Math. Biol. 53, 3–32 - PubMed
LinkOut - more resources
Full Text Sources

