Constitutive α- and β-secretase cleavages of the amyloid precursor protein are partially coupled in neurons, but not in frequently used cell lines
- PMID: 22940630
- PMCID: PMC4310234
- DOI: 10.1016/j.nbd.2012.08.011
Constitutive α- and β-secretase cleavages of the amyloid precursor protein are partially coupled in neurons, but not in frequently used cell lines
Abstract
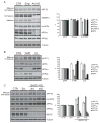
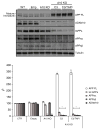
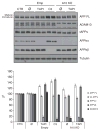
Proteolytic cleavage of the amyloid precursor protein (APP) by the two proteases α- and β-secretases controls the generation of the amyloid β peptide (Aβ), a key player in Alzheimer's disease pathogenesis. The α-secretase ADAM10 and the β-secretase BACE1 have opposite effects on Aβ generation and are assumed to compete for APP as a substrate, such that their cleavages are inversely coupled. This concept was mainly demonstrated in studies using activation or overexpression of α- and β-secretases. Here, we report that this inverse coupling is not seen to the same extent upon inhibition of the endogenous proteases. Genetic and pharmacological inhibition of ADAM10 and BACE1 revealed that the endogenous, constitutive α-secretase cleavage of APP is largely uncoupled from β-secretase cleavage and Aβ generation in neuroglioma H4 cells and in neuronally differentiated SH-SY5Y cells. In contrast, inverse coupling was observed in primary cortical neurons. However, this coupling was not bidirectional. Inhibition of BACE1 increased ADAM10 cleavage of APP, but a reduction of ADAM10 activity did not increase the BACE1 cleavage of APP in the neurons. Our analysis shows that the inverse coupling of the endogenous α- and β-secretase cleavages depends on the cellular model and suggests that a reduction of ADAM10 activity is unlikely to increase the AD risk through increased β-secretase cleavage.
Keywords: Alpha-secretase; Alzheimer's disease; Amyloid precursor protein; Beta-secretase.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Determination of the proteolytic cleavage sites of the amyloid precursor-like protein 2 by the proteases ADAM10, BACE1 and γ-secretase.PLoS One. 2011;6(6):e21337. doi: 10.1371/journal.pone.0021337. Epub 2011 Jun 17. PLoS One. 2011. PMID: 21695060 Free PMC article.
-
ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons.EMBO J. 2010 Sep 1;29(17):3020-32. doi: 10.1038/emboj.2010.167. Epub 2010 Jul 30. EMBO J. 2010. PMID: 20676056 Free PMC article.
-
η-Secretase processing of APP inhibits neuronal activity in the hippocampus.Nature. 2015 Oct 15;526(7573):443-7. doi: 10.1038/nature14864. Epub 2015 Aug 31. Nature. 2015. PMID: 26322584 Free PMC article.
-
Alpha-secretase cleavage of the amyloid precursor protein: proteolysis regulated by signaling pathways and protein trafficking.Curr Alzheimer Res. 2012 Feb;9(2):165-77. doi: 10.2174/156720512799361655. Curr Alzheimer Res. 2012. PMID: 21605033 Review.
-
BACE1: the beta-secretase enzyme in Alzheimer's disease.J Mol Neurosci. 2004;23(1-2):105-14. doi: 10.1385/JMN:23:1-2:105. J Mol Neurosci. 2004. PMID: 15126696 Review.
Cited by
-
The metalloprotease ADAM10 (a disintegrin and metalloprotease 10) undergoes rapid, postlysis autocatalytic degradation.FASEB J. 2018 Jul;32(7):3560-3573. doi: 10.1096/fj.201700823RR. Epub 2018 Feb 7. FASEB J. 2018. PMID: 29430990 Free PMC article.
-
Dietary (-)-epicatechin as a potent inhibitor of βγ-secretase amyloid precursor protein processing.Neurobiol Aging. 2015 Jan;36(1):178-87. doi: 10.1016/j.neurobiolaging.2014.07.032. Epub 2014 Jul 30. Neurobiol Aging. 2015. PMID: 25316600 Free PMC article.
-
Shifting the balance: soluble ADAM10 as a potential treatment for Alzheimer's disease.Front Aging Neurosci. 2023 May 17;15:1171123. doi: 10.3389/fnagi.2023.1171123. eCollection 2023. Front Aging Neurosci. 2023. PMID: 37266401 Free PMC article.
-
A Greek Tragedy: The Growing Complexity of Alzheimer Amyloid Precursor Protein Proteolysis.J Biol Chem. 2016 Sep 9;291(37):19235-44. doi: 10.1074/jbc.R116.746032. Epub 2016 Jul 29. J Biol Chem. 2016. PMID: 27474742 Free PMC article. Review.
-
NrCAM is a marker for substrate-selective activation of ADAM10 in Alzheimer's disease.EMBO Mol Med. 2019 Apr;11(4):e9695. doi: 10.15252/emmm.201809695. EMBO Mol Med. 2019. PMID: 30833305 Free PMC article.
References
-
- Ahmad M, Takino T, Miyamori H, Yoshizaki T, Furukawa M, Sato H. Cleavage of amyloid-beta precursor protein (APP) by membrane-type matrix metalloproteinases. J Biochem. 2006 Mar;139(3):517–526. - PubMed
-
- Amtul Z, Wang L, Westaway D, Rozmahel RF. Neuroprotective mechanism conferred by 17beta-estradiol on the biochemical basis of Alzheimer’s disease. Neuroscience. 2010 Aug 25;169(2):781–786. Epub 2010 May 21. - PubMed
-
- Caccamo A, Oddo S, Billings LM, Green KN, Martinez-Coria H, Fisher A, LaFerla FM. Neuron. 2006 Mar 2;49(5):671–682. - PubMed
-
- Colombo A, Repici M, Pesaresi M, Santambrogio S, Forloni G, Borsello T. The TAT-JNK inhibitor peptide interferes with beta amyloid protein stability. Cell Death Differ. 2007 Oct;14(10):1845–1848. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

