Hsp70 protein complexes as drug targets
- PMID: 22920901
- PMCID: PMC3593251
- DOI: 10.2174/138161213804143699
Hsp70 protein complexes as drug targets
Abstract
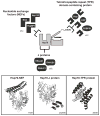
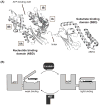
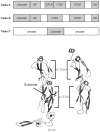
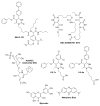
Heat shock protein 70 (Hsp70) plays critical roles in proteostasis and is an emerging target for multiple diseases. However, competitive inhibition of the enzymatic activity of Hsp70 has proven challenging and, in some cases, may not be the most productive way to redirect Hsp70 function. Another approach is to inhibit Hsp70's interactions with important co-chaperones, such as J proteins, nucleotide exchange factors (NEFs) and tetratricopeptide repeat (TPR) domain-containing proteins. These co-chaperones normally bind Hsp70 and guide its many diverse cellular activities. Complexes between Hsp70 and co-chaperones have been shown to have specific functions, including roles in pro-folding, pro-degradation and pro-trafficking pathways. Thus, a promising strategy may be to block protein- protein interactions between Hsp70 and its co-chaperones or to target allosteric sites that disrupt these contacts. Such an approach might shift the balance of Hsp70 complexes and re-shape the proteome and it has the potential to restore healthy proteostasis. In this review, we discuss specific challenges and opportunities related to these goals. By pursuing Hsp70 complexes as drug targets, we might not only develop new leads for therapeutic development, but also discover new chemical probes for use in understanding Hsp70 biology.
Figures

Similar articles
-
Multivalent protein-protein interactions are pivotal regulators of eukaryotic Hsp70 complexes.Cell Stress Chaperones. 2022 Jul;27(4):397-415. doi: 10.1007/s12192-022-01281-1. Epub 2022 Jun 7. Cell Stress Chaperones. 2022. PMID: 35670950 Free PMC article. Review.
-
High-throughput screen for inhibitors of protein-protein interactions in a reconstituted heat shock protein 70 (Hsp70) complex.J Biol Chem. 2018 Mar 16;293(11):4014-4025. doi: 10.1074/jbc.RA117.001575. Epub 2018 Feb 2. J Biol Chem. 2018. PMID: 29414793 Free PMC article.
-
Pharmacological targeting of the Hsp70 chaperone.Curr Top Med Chem. 2009;9(15):1337-51. doi: 10.2174/156802609789895674. Curr Top Med Chem. 2009. PMID: 19860737 Free PMC article. Review.
-
Cytosolic protein quality control machinery: Interactions of Hsp70 with a network of co-chaperones and substrates.Exp Biol Med (Maywood). 2021 Jun;246(12):1419-1434. doi: 10.1177/1535370221999812. Epub 2021 Mar 17. Exp Biol Med (Maywood). 2021. PMID: 33730888 Free PMC article. Review.
-
Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro.J Biol Chem. 2014 Jan 17;289(3):1402-14. doi: 10.1074/jbc.M113.521997. Epub 2013 Dec 5. J Biol Chem. 2014. PMID: 24318877 Free PMC article.
Cited by
-
Heat Shock Protein 70 as a Double Agent Acting Inside and Outside the Cell: Insights into Autoimmunity.Int J Mol Sci. 2020 Jul 26;21(15):5298. doi: 10.3390/ijms21155298. Int J Mol Sci. 2020. PMID: 32722570 Free PMC article. Review.
-
Tackling Sleeping Sickness: Current and Promising Therapeutics and Treatment Strategies.Int J Mol Sci. 2023 Aug 7;24(15):12529. doi: 10.3390/ijms241512529. Int J Mol Sci. 2023. PMID: 37569903 Free PMC article. Review.
-
Hsp70 molecular chaperones: multifunctional allosteric holding and unfolding machines.Biochem J. 2019 Jun 14;476(11):1653-1677. doi: 10.1042/BCJ20170380. Biochem J. 2019. PMID: 31201219 Free PMC article. Review.
-
Analogs of the Allosteric Heat Shock Protein 70 (Hsp70) Inhibitor, MKT-077, as Anti-Cancer Agents.ACS Med Chem Lett. 2013 Nov 14;4(11):1042-7. doi: 10.1021/ml400204n. ACS Med Chem Lett. 2013. PMID: 24312699 Free PMC article.
-
The hop-like stress-induced protein 1 cochaperone is a novel cell-intrinsic restriction factor for mitochondrial tombusvirus replication.J Virol. 2014 Aug;88(16):9361-78. doi: 10.1128/JVI.00561-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920799 Free PMC article.
References
-
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–51. - PubMed
-
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Ann Rev Biochem. 2001;70:603–47. - PubMed
-
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. - PubMed
-
- Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–10. - PubMed

