Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1
- PMID: 22916024
- PMCID: PMC3415433
- DOI: 10.1371/journal.pgen.1002855
Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1
Abstract
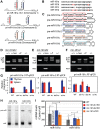
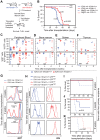
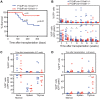
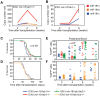
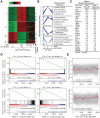
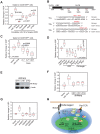
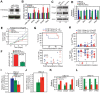
Oncogenes, which are essential for tumor initiation, development, and maintenance, are valuable targets for cancer therapy. However, it remains a challenge to effectively inhibit oncogene activity by targeting their downstream pathways without causing significant toxicity to normal tissues. Here we show that deletion of mir-181a-1/b-1 expression inhibits the development of Notch1 oncogene-induced T cell acute lymphoblastic leukemia (T-ALL). mir-181a-1/b-1 controls the strength and threshold of Notch activity in tumorigenesis in part by dampening multiple negative feedback regulators downstream of NOTCH and pre-T cell receptor (TCR) signaling pathways. Importantly, although Notch oncogenes utilize normal thymic progenitor cell genetic programs for tumor transformation, comparative analyses of mir-181a-1/b-1 function in normal thymocyte and tumor development demonstrate that mir-181a-1/b-1 can be specifically targeted to inhibit tumor development with little toxicity to normal development. Finally, we demonstrate that mir-181a-1/b-1, but not mir-181a-2b-2 and mir-181-c/d, controls the development of normal thymic T cells and leukemia cells. Together, these results illustrate that NOTCH oncogene activity in tumor development can be selectively inhibited by targeting the molecular networks controlled by mir-181a-1/b-1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A microRNA-mediated regulatory loop modulates NOTCH and MYC oncogenic signals in B- and T-cell malignancies.Leukemia. 2015 Apr;29(4):968-76. doi: 10.1038/leu.2014.302. Epub 2014 Oct 14. Leukemia. 2015. PMID: 25311243 Free PMC article.
-
Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150.Blood. 2011 Jun 30;117(26):7053-62. doi: 10.1182/blood-2010-12-326629. Epub 2011 May 6. Blood. 2011. PMID: 21551231
-
Regulation of miR-181a expression in T cell aging.Nat Commun. 2018 Aug 3;9(1):3060. doi: 10.1038/s41467-018-05552-3. Nat Commun. 2018. PMID: 30076309 Free PMC article.
-
Oncogenic Notch signaling in T-cell and B-cell lymphoproliferative disorders.Curr Opin Hematol. 2016 Jul;23(4):362-70. doi: 10.1097/MOH.0000000000000254. Curr Opin Hematol. 2016. PMID: 27135981 Free PMC article. Review.
-
Notch and Wnt signaling in T-lymphocyte development and acute lymphoblastic leukemia.Leukemia. 2006 Jul;20(7):1197-205. doi: 10.1038/sj.leu.2404255. Epub 2006 May 11. Leukemia. 2006. PMID: 16688226 Review.
Cited by
-
The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis.Immunity. 2013 May 23;38(5):984-97. doi: 10.1016/j.immuni.2013.02.021. Epub 2013 Apr 25. Immunity. 2013. PMID: 23623381 Free PMC article.
-
Outside the coding genome, mammalian microRNAs confer structural and functional complexity.Sci Signal. 2015 Mar 17;8(368):re2. doi: 10.1126/scisignal.2005813. Sci Signal. 2015. PMID: 25783159 Free PMC article. Review.
-
Attenuation of antigenic immunogenicity by kynurenine, a novel suppressive adjuvant.Hum Vaccin Immunother. 2014;10(5):1295-305. doi: 10.4161/hv.28099. Epub 2014 Feb 28. Hum Vaccin Immunother. 2014. PMID: 24583631 Free PMC article.
-
Integrins and the Metastasis-like Dissemination of Acute Lymphoblastic Leukemia to the Central Nervous System.Cancers (Basel). 2023 Apr 27;15(9):2504. doi: 10.3390/cancers15092504. Cancers (Basel). 2023. PMID: 37173970 Free PMC article. Review.
-
Differential Co-expression and Regulatory Network Analysis Uncover the Relapse Factor and Mechanism of T Cell Acute Leukemia.Mol Ther Nucleic Acids. 2018 Sep 7;12:184-194. doi: 10.1016/j.omtn.2018.05.003. Epub 2018 May 29. Mol Ther Nucleic Acids. 2018. PMID: 30195757 Free PMC article.
References
-
- Medina PP, Nolde M, Slack FJ (2010) OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467: 86–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

