Oxa1-ribosome complexes coordinate the assembly of cytochrome C oxidase in mitochondria
- PMID: 22904327
- PMCID: PMC3464553
- DOI: 10.1074/jbc.M112.382630
Oxa1-ribosome complexes coordinate the assembly of cytochrome C oxidase in mitochondria
Abstract
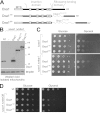
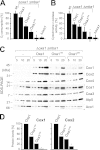
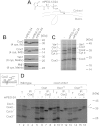
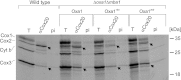
The terminal enzyme of the respiratory chain, cytochrome c oxidase, consists of a hydrophobic reaction center formed by three mitochondrially encoded subunits with which 9-10 nuclear encoded subunits are associated. The three core subunits are synthesized on mitochondrial ribosomes and inserted into the inner membrane in a co-translational reaction facilitated by the Oxa1 insertase. Oxa1 consists of an N-terminal insertase domain and a C-terminal ribosome-binding region. Mutants lacking the C-terminal region show specific defects in co-translational insertion, suggesting that the close contact of the ribosome with the insertase promotes co-translational insertion of nascent chains. In this study, we inserted flexible linkers of 100 or 200 amino acid residues between the insertase domain and ribosome-binding region of Oxa1 of Saccharomyces cerevisiae. In the absence of the ribosome receptor Mba1, these linkers caused a length-dependent decrease in mitochondrial respiratory activity caused by diminished levels of cytochrome c oxidase. Interestingly, considerable amounts of mitochondrial translation products were still integrated into the inner membrane in these linker mutants. However, they showed severe defects in later stages of the biogenesis process, presumably during assembly into functional complexes. Our observations suggest that the close proximity of Oxa1 to ribosomes is not only used to improve membrane insertion but is also critical for the productive assembly of the subunits of the cytochrome c oxidase. This points to a role for Oxa1 in the spatial coordination of the ribosome with assembly factors that are critical for enzyme biogenesis.
Figures


Similar articles
-
Mba1, a membrane-associated ribosome receptor in mitochondria.EMBO J. 2006 Apr 19;25(8):1603-10. doi: 10.1038/sj.emboj.7601070. Epub 2006 Apr 6. EMBO J. 2006. PMID: 16601683 Free PMC article.
-
Translocation and assembly of mitochondrially coded Saccharomyces cerevisiae cytochrome c oxidase subunit Cox2 by Oxa1 and Yme1 in the absence of Cox18.Genetics. 2009 Jun;182(2):519-28. doi: 10.1534/genetics.109.101196. Epub 2009 Mar 23. Genetics. 2009. PMID: 19307606 Free PMC article.
-
Mapping of the Saccharomyces cerevisiae Oxa1-mitochondrial ribosome interface and identification of MrpL40, a ribosomal protein in close proximity to Oxa1 and critical for oxidative phosphorylation complex assembly.Eukaryot Cell. 2009 Nov;8(11):1792-802. doi: 10.1128/EC.00219-09. Epub 2009 Sep 25. Eukaryot Cell. 2009. PMID: 19783770 Free PMC article.
-
Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes.Biochim Biophys Acta. 2009 Jan;1793(1):60-70. doi: 10.1016/j.bbamcr.2008.05.004. Epub 2008 May 15. Biochim Biophys Acta. 2009. PMID: 18522806 Free PMC article. Review.
-
Co-translational membrane insertion of mitochondrially encoded proteins.Biochim Biophys Acta. 2010 Jun;1803(6):767-75. doi: 10.1016/j.bbamcr.2009.11.010. Epub 2009 Dec 2. Biochim Biophys Acta. 2010. PMID: 19962410 Review.
Cited by
-
Types and Functions of Mitoribosome-Specific Ribosomal Proteins across Eukaryotes.Int J Mol Sci. 2022 Mar 23;23(7):3474. doi: 10.3390/ijms23073474. Int J Mol Sci. 2022. PMID: 35408834 Free PMC article. Review.
-
Protein insertion into the inner membrane of mitochondria: routes and mechanisms.FEBS Open Bio. 2024 Oct;14(10):1627-1639. doi: 10.1002/2211-5463.13806. Epub 2024 Apr 25. FEBS Open Bio. 2024. PMID: 38664330 Free PMC article. Review.
-
Ribosome-Associated Mba1 Escorts Cox2 from Insertion Machinery to Maturing Assembly Intermediates.Mol Cell Biol. 2016 Oct 28;36(22):2782-2793. doi: 10.1128/MCB.00361-16. Print 2016 Nov 15. Mol Cell Biol. 2016. PMID: 27550809 Free PMC article.
-
Chloroplast Ribosomes Interact With the Insertase Alb3 in the Thylakoid Membrane.Front Plant Sci. 2021 Dec 23;12:781857. doi: 10.3389/fpls.2021.781857. eCollection 2021. Front Plant Sci. 2021. PMID: 35003166 Free PMC article.
-
OXA2b is Crucial for Proper Membrane Insertion of COX2 during Biogenesis of Complex IV in Plant Mitochondria.Plant Physiol. 2019 Feb;179(2):601-615. doi: 10.1104/pp.18.01286. Epub 2018 Nov 28. Plant Physiol. 2019. PMID: 30487140 Free PMC article.
References
-
- Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 - PubMed
-
- Kramer G., Boehringer D., Ban N., Bukau B. (2009) The ribosome as a platform for co-translational processing, folding, and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 - PubMed
-
- Giglione C., Fieulaine S., Meinnel T. (2009) Co-translational processing mechanisms: towards a dynamic 3D model. Trends Biochem. Sci. 34, 417–426 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

