SUMO, a heavyweight player in plant abiotic stress responses
- PMID: 22903295
- PMCID: PMC11114757
- DOI: 10.1007/s00018-012-1094-2
SUMO, a heavyweight player in plant abiotic stress responses
Abstract
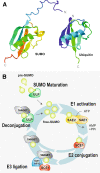
Protein post-translational modifications diversify the proteome and install new regulatory levels that are crucial for the maintenance of cellular homeostasis. Over the last decade, the ubiquitin-like modifying peptide small ubiquitin-like modifier (SUMO) has been shown to regulate various nuclear processes, including transcriptional control. In plants, the sumoylation pathway has been significantly implicated in the response to environmental stimuli, including heat, cold, drought, and salt stresses, modulation of abscisic acid and other hormones, and nutrient homeostasis. This review focuses on the emerging importance of SUMO in the abiotic stress response, summarizing the molecular implications of sumoylation and emphasizing how high-throughput approaches aimed at identifying the full set of SUMO targets will greatly enhance our understanding of the SUMO-abiotic stress association.
Figures


Similar articles
-
Small ubiquitin-like modifiers E3 ligases in plant stress.Funct Plant Biol. 2024 Apr;51:FP24032. doi: 10.1071/FP24032. Funct Plant Biol. 2024. PMID: 38669463 Review.
-
SUMO and SUMOylation in plants.Mol Cells. 2011 Oct;32(4):305-16. doi: 10.1007/s10059-011-0122-7. Epub 2011 Sep 9. Mol Cells. 2011. PMID: 21912873 Free PMC article. Review.
-
A stress inducible SUMO conjugating enzyme gene (SaSce9) from a grass halophyte Spartina alterniflora enhances salinity and drought stress tolerance in Arabidopsis.BMC Plant Biol. 2012 Oct 10;12:187. doi: 10.1186/1471-2229-12-187. BMC Plant Biol. 2012. PMID: 23051937 Free PMC article.
-
Regulation of Plant Cellular and Organismal Development by SUMO.Adv Exp Med Biol. 2017;963:227-247. doi: 10.1007/978-3-319-50044-7_14. Adv Exp Med Biol. 2017. PMID: 28197916 Review.
-
Understanding SUMO-mediated adaptive responses in plants to improve crop productivity.Essays Biochem. 2022 Aug 5;66(2):155-168. doi: 10.1042/EBC20210068. Essays Biochem. 2022. PMID: 35920279 Free PMC article. Review.
Cited by
-
ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress.Int J Mol Sci. 2022 Jan 28;23(3):1549. doi: 10.3390/ijms23031549. Int J Mol Sci. 2022. PMID: 35163471 Free PMC article. Review.
-
SUMO E3 ligase SIZ1 connects sumoylation and reactive oxygen species homeostasis processes in Arabidopsis.Plant Physiol. 2022 Jun 1;189(2):934-954. doi: 10.1093/plphys/kiac085. Plant Physiol. 2022. PMID: 35238389 Free PMC article.
-
FLC-mediated flowering repression is positively regulated by sumoylation.J Exp Bot. 2014 Jan;65(1):339-51. doi: 10.1093/jxb/ert383. Epub 2013 Nov 11. J Exp Bot. 2014. PMID: 24218331 Free PMC article.
-
SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2015 Sep 1;112(35):11108-13. doi: 10.1073/pnas.1415260112. Epub 2015 Aug 17. Proc Natl Acad Sci U S A. 2015. PMID: 26283376 Free PMC article.
-
Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases.Front Plant Sci. 2014 May 20;5:190. doi: 10.3389/fpls.2014.00190. eCollection 2014. Front Plant Sci. 2014. PMID: 24904600 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

