Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses
- PMID: 22901812
- PMCID: PMC3499626
- DOI: 10.1016/j.cell.2012.05.049
Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses
Abstract
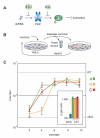
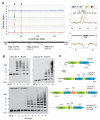
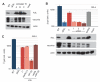
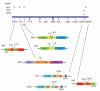
In contrast to RNA viruses, double-stranded DNA viruses have low mutation rates yet must still adapt rapidly in response to changing host defenses. To determine mechanisms of adaptation, we subjected the model poxvirus vaccinia to serial propagation in human cells, where its antihost factor K3L is maladapted against the antiviral protein kinase R (PKR). Viruses rapidly acquired higher fitness via recurrent K3L gene amplifications, incurring up to 7%-10% increases in genome size. These transient gene expansions were necessary and sufficient to counteract human PKR and facilitated the gain of an adaptive amino acid substitution in K3L that also defeats PKR. Subsequent reductions in gene amplifications offset the costs associated with larger genome size while retaining adaptive substitutions. Our discovery of viral "gene-accordions" explains how poxviruses can rapidly adapt to defeat different host defenses despite low mutation rates and reveals how classical Red Queen conflicts can progress through unrecognized intermediates.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Poxvirus use a "gene accordion" to tune out host defenses.Cell. 2012 Aug 17;150(4):671-2. doi: 10.1016/j.cell.2012.07.026. Cell. 2012. PMID: 22901801
-
Viral evolution: Accordion adaptations go viral.Nat Rev Microbiol. 2012 Oct;10(10):669. doi: 10.1038/nrmicro2883. Epub 2012 Sep 10. Nat Rev Microbiol. 2012. PMID: 22961338 No abstract available.
Similar articles
-
Emergence of a Viral RNA Polymerase Variant during Gene Copy Number Amplification Promotes Rapid Evolution of Vaccinia Virus.J Virol. 2017 Jan 31;91(4):e01428-16. doi: 10.1128/JVI.01428-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27928012 Free PMC article.
-
Systematic genetic characterization of the human PKR kinase domain highlights its functional malleability to escape a poxvirus substrate mimic.Elife. 2024 Nov 12;13:RP99575. doi: 10.7554/eLife.99575. Elife. 2024. PMID: 39531012 Free PMC article.
-
Protein kinase R reveals an evolutionary model for defeating viral mimicry.Nature. 2009 Jan 22;457(7228):485-9. doi: 10.1038/nature07529. Epub 2008 Nov 30. Nature. 2009. PMID: 19043403 Free PMC article.
-
[Progress on host range factors and their mechanisms of poxvirus].Bing Du Xue Bao. 2013 Nov;29(6):655-61. Bing Du Xue Bao. 2013. PMID: 24520773 Review. Chinese.
-
Poxviruses and the evolution of host range and virulence.Infect Genet Evol. 2014 Jan;21:15-40. doi: 10.1016/j.meegid.2013.10.014. Epub 2013 Oct 24. Infect Genet Evol. 2014. PMID: 24161410 Free PMC article. Review.
Cited by
-
New insights into the evolution of Entomopoxvirinae from the complete genome sequences of four entomopoxviruses infecting Adoxophyes honmai, Choristoneura biennis, Choristoneura rosaceana, and Mythimna separata.J Virol. 2013 Jul;87(14):7992-8003. doi: 10.1128/JVI.00453-13. Epub 2013 May 15. J Virol. 2013. PMID: 23678178 Free PMC article.
-
Inactivation of Genes by Frameshift Mutations Provides Rapid Adaptation of an Attenuated Vaccinia Virus.J Virol. 2020 Aug 31;94(18):e01053-20. doi: 10.1128/JVI.01053-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32669330 Free PMC article.
-
Molecular detection of monkeypox and related viruses: challenges and opportunities.Virus Genes. 2023 Jun;59(3):343-350. doi: 10.1007/s11262-023-01975-3. Epub 2023 Feb 6. Virus Genes. 2023. PMID: 36746846 Free PMC article. Review.
-
The Role of Ku70 as a Cytosolic DNA Sensor in Innate Immunity and Beyond.Front Cell Infect Microbiol. 2021 Oct 21;11:761983. doi: 10.3389/fcimb.2021.761983. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34746031 Free PMC article. Review.
-
The prospective outcome of the monkeypox outbreak in 2022 and characterization of monkeypox disease immunobiology.Front Cell Infect Microbiol. 2023 Jul 18;13:1196699. doi: 10.3389/fcimb.2023.1196699. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37533932 Free PMC article.
References
-
- Andersson DI, Slechta ES, Roth JR. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

