Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7)
- PMID: 22898984
- PMCID: PMC3446710
- DOI: 10.1261/rna.034538.112
Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7)
Abstract
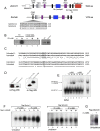
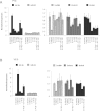
The pluripotency factor Lin28 recruits a 3' terminal uridylyl transferase (TUTase) to selectively block let-7 microRNA biogenesis in undifferentiated cells. Zcchc11 (TUTase4/TUT4) was previously identified as an enzyme responsible for Lin28-mediated pre-let-7 uridylation and control of let-7 expression. Here we investigate the protein and RNA determinants for this interaction. Biochemical dissection and reconstitution assays reveal the TUTase domains necessary and sufficient for Lin28-enhanced pre-let-7 uridylation. A single C2H2-type zinc finger domain of Zcchc11 was found to be responsible for the functional interaction with Lin28. We identify Zcchc6 (TUTase7) as an alternative TUTase that functions with Lin28 in vitro, and accordingly, we find Zcchc11 and Zcchc6 redundantly control let-7 biogenesis in embryonic stem cells. Our study indicates that Lin28 uses two different TUTases to control let-7 expression and has important implications for stem cell biology as well as cancer.
Figures


Comment in
-
Small RNAs: controlling maturity.Nat Rev Mol Cell Biol. 2012 Dec;13(12):753. doi: 10.1038/nrm3475. Epub 2012 Nov 15. Nat Rev Mol Cell Biol. 2012. PMID: 23151665 No abstract available.
Similar articles
-
Identification of small molecule inhibitors of Zcchc11 TUTase activity.RNA Biol. 2015;12(8):792-800. doi: 10.1080/15476286.2015.1058478. RNA Biol. 2015. PMID: 26114892 Free PMC article.
-
Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells.Nat Struct Mol Biol. 2009 Oct;16(10):1021-5. doi: 10.1038/nsmb.1676. Epub 2009 Aug 27. Nat Struct Mol Biol. 2009. PMID: 19713958 Free PMC article.
-
Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4).Nucleic Acids Res. 2014 Oct;42(18):11777-91. doi: 10.1093/nar/gku805. Epub 2014 Sep 15. Nucleic Acids Res. 2014. PMID: 25223788 Free PMC article.
-
3' RNA Uridylation in Epitranscriptomics, Gene Regulation, and Disease.Front Mol Biosci. 2018 Jul 13;5:61. doi: 10.3389/fmolb.2018.00061. eCollection 2018. Front Mol Biosci. 2018. PMID: 30057901 Free PMC article. Review.
-
The LIN28/let-7 Pathway in Cancer.Front Genet. 2017 Mar 28;8:31. doi: 10.3389/fgene.2017.00031. eCollection 2017. Front Genet. 2017. PMID: 28400788 Free PMC article. Review.
Cited by
-
MicroRNAs in Control of Stem Cells in Normal and Malignant Hematopoiesis.Curr Stem Cell Rep. 2016 Sep;2(3):183-196. doi: 10.1007/s40778-016-0057-1. Epub 2016 Jul 1. Curr Stem Cell Rep. 2016. PMID: 27547713 Free PMC article.
-
Selective Suppression of the Splicing-Mediated MicroRNA Pathway by the Terminal Uridyltransferase Tailor.Mol Cell. 2015 Jul 16;59(2):217-28. doi: 10.1016/j.molcel.2015.05.034. Epub 2015 Jul 2. Mol Cell. 2015. PMID: 26145174 Free PMC article.
-
Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.Nucleic Acids Res. 2015 Mar 11;43(5):2968-79. doi: 10.1093/nar/gkv122. Epub 2015 Feb 20. Nucleic Acids Res. 2015. PMID: 25712096 Free PMC article.
-
Genetic Inactivation of ZCCHC6 Suppresses Interleukin-6 Expression and Reduces the Severity of Experimental Osteoarthritis in Mice.Arthritis Rheumatol. 2019 Apr;71(4):583-593. doi: 10.1002/art.40751. Epub 2019 Mar 6. Arthritis Rheumatol. 2019. PMID: 30302948 Free PMC article.
-
Terminal nucleotidyl transferases (TENTs) in mammalian RNA metabolism.Philos Trans R Soc Lond B Biol Sci. 2018 Nov 5;373(1762):20180162. doi: 10.1098/rstb.2018.0162. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 30397099 Free PMC article. Review.
References
-
- Ambros V, Horvitz HR 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226: 409–416 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
