Protein folding rates and thermodynamic stability are key determinants for interaction with the Hsp70 chaperone system
- PMID: 22886941
- PMCID: PMC3526991
- DOI: 10.1002/pro.2139
Protein folding rates and thermodynamic stability are key determinants for interaction with the Hsp70 chaperone system
Abstract
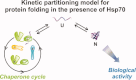
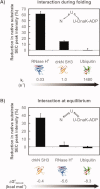
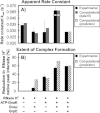
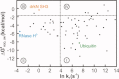
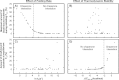
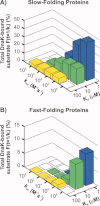
The Hsp70 family of molecular chaperones participates in vital cellular processes including the heat shock response and protein homeostasis. E. coli's Hsp70, known as DnaK, works in concert with the DnaJ and GrpE co-chaperones (K/J/E chaperone system), and mediates cotranslational and post-translational protein folding in the cytoplasm. While the role of the K/J/E chaperones is well understood in the presence of large substrates unable to fold independently, it is not known if and how K/J/E modulates the folding of smaller proteins able to fold even in the absence of chaperones. Here, we combine experiments and computation to evaluate the significance of kinetic partitioning as a model to describe the interplay between protein folding and binding to the K/J/E chaperone system. First, we target three nonobligatory substrates, that is, proteins that do not require chaperones to fold. The experimentally observed chaperone association of these client proteins during folding is entirely consistent with predictions from kinetic partitioning. Next, we develop and validate a computational model (CHAMP70) that assumes kinetic partitioning of substrates between folding and interaction with K/J/E. CHAMP70 quantitatively predicts the experimentally measured interaction of RNase H(D) as it refolds in the presence of various chaperones. CHAMP70 shows that substrates are posed to interact with K/J/E only if they are slow-folding proteins with a folding rate constant k(f) <50 s⁻¹, and/or thermodynamically unstable proteins with a folding free energy ΔG⁰ (UN) ≥-2 kcal mol⁻¹. Hence, the K/J/E system is tuned to use specific protein folding rates and thermodynamic stabilities as substrate selection criteria.
Copyright © 2012 The Protein Society.
Figures


Similar articles
-
Transient interactions of a slow-folding protein with the Hsp70 chaperone machinery.Protein Sci. 2012 Jul;21(7):1042-55. doi: 10.1002/pro.2087. Epub 2012 Jun 11. Protein Sci. 2012. PMID: 22549943 Free PMC article.
-
Regulation of ATPase and chaperone cycle of DnaK from Thermus thermophilus by the nucleotide exchange factor GrpE.J Mol Biol. 2001 Feb 2;305(5):1173-83. doi: 10.1006/jmbi.2000.4373. J Mol Biol. 2001. PMID: 11162122
-
Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein.Nat Commun. 2020 Jan 17;11(1):365. doi: 10.1038/s41467-019-14245-4. Nat Commun. 2020. PMID: 31953415 Free PMC article.
-
Structural and Biochemical Properties of Hsp40/Hsp70 Chaperone System.Adv Exp Med Biol. 2020;1243:3-20. doi: 10.1007/978-3-030-40204-4_1. Adv Exp Med Biol. 2020. PMID: 32297208 Review.
-
Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions.Protein Sci. 2005 Jul;14(7):1697-709. doi: 10.1110/ps.051406805. Protein Sci. 2005. PMID: 15987899 Free PMC article. Review.
Cited by
-
Cytosolic protein quality control machinery: Interactions of Hsp70 with a network of co-chaperones and substrates.Exp Biol Med (Maywood). 2021 Jun;246(12):1419-1434. doi: 10.1177/1535370221999812. Epub 2021 Mar 17. Exp Biol Med (Maywood). 2021. PMID: 33730888 Free PMC article. Review.
-
Distribution and solvent exposure of Hsp70 chaperone binding sites across the Escherichia coli proteome.Proteins. 2023 May;91(5):665-678. doi: 10.1002/prot.26456. Epub 2023 Jan 1. Proteins. 2023. PMID: 36539330 Free PMC article.
-
Protein folding in vitro and in the cell: From a solitary journey to a team effort.Biophys Chem. 2022 Aug;287:106821. doi: 10.1016/j.bpc.2022.106821. Epub 2022 Apr 29. Biophys Chem. 2022. PMID: 35667131 Free PMC article. Review.
-
KLR-70: A Novel Cationic Inhibitor of the Bacterial Hsp70 Chaperone.Biochemistry. 2020 May 26;59(20):1946-1960. doi: 10.1021/acs.biochem.0c00320. Epub 2020 May 4. Biochemistry. 2020. PMID: 32326704 Free PMC article.
-
DnaK response to expression of protein mutants is dependent on translation rate and stability.Commun Biol. 2022 Jun 16;5(1):597. doi: 10.1038/s42003-022-03542-2. Commun Biol. 2022. PMID: 35710941 Free PMC article.
References
-
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. - PubMed
-
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Ann Rev Biochem. 2001;70:603–647. - PubMed
-
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852. - PubMed
-
- Mayer MP, Rudiger S, Bukau B. Molecular basis for interactions of the DnaK chaperone with substrates. Biol Chem. 2000;381:877–885. - PubMed
-
- Chesnokova LS, Slepenkov SV, Protasevich II, Sehorn MG, Brouillette CG, Witt SN. Deletion of DnaK's lid strengthens binding to the nucleotide exchange factor, GrpE: a kinetic and thermodynamic analysis. Biochemistry. 2003;42:9028–9040. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

