Patterns of DNA methylation in development, division of labor and hybridization in an ant with genetic caste determination
- PMID: 22879983
- PMCID: PMC3411777
- DOI: 10.1371/journal.pone.0042433
Patterns of DNA methylation in development, division of labor and hybridization in an ant with genetic caste determination
Abstract
Background: DNA methylation is a common regulator of gene expression, including acting as a regulator of developmental events and behavioral changes in adults. Using the unique system of genetic caste determination in Pogonomyrmex barbatus, we were able to document changes in DNA methylation during development, and also across both ancient and contemporary hybridization events.
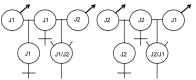
Methodology/principal findings: Sodium bisulfite sequencing demonstrated in vivo methylation of symmetric CG dinucleotides in P. barbatus. We also found methylation of non-CpG sequences. This validated two bioinformatics methods for predicting gene methylation, the bias in observed to expected ratio of CpG dinucleotides and the density of CpG/TpG single nucleotide polymorphisms (SNP). Frequencies of genomic DNA methylation were determined for different developmental stages and castes using ms-AFLP assays. The genetic caste determination system (GCD) is probably the product of an ancestral hybridization event between P. barbatus and P. rugosus. Two lineages obligately co-occur within a GCD population, and queens are derived from intra-lineage matings whereas workers are produced from inter-lineage matings. Relative DNA methylation levels of queens and workers from GCD lineages (contemporary hybrids) were not significantly different until adulthood. Virgin queens had significantly higher relative levels of DNA methylation compared to workers. Worker DNA methylation did not vary among developmental stages within each lineage, but was significantly different between the currently hybridizing lineages. Finally, workers of the two genetic caste determination lineages had half as many methylated cytosines as workers from the putative parental species, which have environmental caste determination.
Conclusions/significance: These results suggest that DNA methylation may be a conserved regulatory mechanism moderating division of labor in both bees and ants. Current and historic hybridization appear to have altered genomic methylation levels suggesting a possible link between changes in overall DNA methylation and the origin and regulation of genetic caste determination in P. barbatus.
Conflict of interest statement
Figures

Similar articles
-
The role of epigenetics, particularly DNA methylation, in the evolution of caste in insect societies.Philos Trans R Soc Lond B Biol Sci. 2021 Jun 7;376(1826):20200115. doi: 10.1098/rstb.2020.0115. Epub 2021 Apr 19. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 33866805 Free PMC article. Review.
-
Characterization and distribution of Pogonomyrmex harvester ant lineages with genetic caste determination.Mol Ecol. 2007 Jan;16(2):367-87. doi: 10.1111/j.1365-294X.2006.03124.x. Mol Ecol. 2007. PMID: 17217351
-
Distribution and evolution of genetic caste determination in Pogonomyrmex seed-harvester ants.Ecology. 2006 Sep;87(9):2171-84. doi: 10.1890/0012-9658(2006)87[2171:daeogc]2.0.co;2. Ecology. 2006. PMID: 16995616
-
Population-wide lineage frequencies predict genetic load in the seed-harvester ant Pogonomyrmex.Proc Natl Acad Sci U S A. 2006 Sep 5;103(36):13433-8. doi: 10.1073/pnas.0606055103. Epub 2006 Aug 28. Proc Natl Acad Sci U S A. 2006. PMID: 16938859 Free PMC article.
-
Genetic caste determination in harvester ants: possible origin and maintenance by cyto-nuclear epistasis.Ecology. 2006 Sep;87(9):2185-93. doi: 10.1890/0012-9658(2006)87[2185:gcdiha]2.0.co;2. Ecology. 2006. PMID: 16995617 Review.
Cited by
-
Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality.Genome Res. 2013 Aug;23(8):1235-47. doi: 10.1101/gr.155408.113. Epub 2013 May 1. Genome Res. 2013. PMID: 23636946 Free PMC article.
-
Effects of ploidy and sex-locus genotype on gene expression patterns in the fire ant Solenopsis invicta.Proc Biol Sci. 2014 Dec 22;281(1797):20141776. doi: 10.1098/rspb.2014.1776. Proc Biol Sci. 2014. PMID: 25355475 Free PMC article.
-
The role of epigenetics, particularly DNA methylation, in the evolution of caste in insect societies.Philos Trans R Soc Lond B Biol Sci. 2021 Jun 7;376(1826):20200115. doi: 10.1098/rstb.2020.0115. Epub 2021 Apr 19. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 33866805 Free PMC article. Review.
-
The draft genome of a socially polymorphic halictid bee, Lasioglossum albipes.Genome Biol. 2013 Dec 20;14(12):R142. doi: 10.1186/gb-2013-14-12-r142. Genome Biol. 2013. PMID: 24359881 Free PMC article.
-
Sexual dimorphism as a facilitator of worker caste evolution in ants.Ecol Evol. 2023 Feb 15;13(2):e9825. doi: 10.1002/ece3.9825. eCollection 2023 Feb. Ecol Evol. 2023. PMID: 36818531 Free PMC article.
References
-
- Razin A, Shemer R (1995) DNA methylation in early development. Hum Mol Genet 4: 1751–1755. - PubMed
-
- Field LM, Lyko F, Mandrioli M, Prantera G (2004) DNA methylation in insects. Insect Mol Biol13: 109–115. - PubMed
-
- Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514. - PubMed
-
- Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919. - PubMed
-
- Xiang H, Zhu J, Chen Q, Dai F, Li X, et al. (2010) Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol 28: 516–520. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

