Highly conserved protective epitopes on influenza B viruses
- PMID: 22878502
- PMCID: PMC3538841
- DOI: 10.1126/science.1222908
Highly conserved protective epitopes on influenza B viruses
Abstract
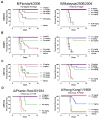
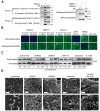
Identification of broadly neutralizing antibodies against influenza A viruses has raised hopes for the development of monoclonal antibody-based immunotherapy and "universal" vaccines for influenza. However, a substantial part of the annual flu burden is caused by two cocirculating, antigenically distinct lineages of influenza B viruses. Here, we report human monoclonal antibodies, CR8033, CR8071, and CR9114, that protect mice against lethal challenge from both lineages. Antibodies CR8033 and CR8071 recognize distinct conserved epitopes in the head region of the influenza B hemagglutinin (HA), whereas CR9114 binds a conserved epitope in the HA stem and protects against lethal challenge with influenza A and B viruses. These antibodies may inform on development of monoclonal antibody-based treatments and a universal flu vaccine for all influenza A and B viruses.
Figures


Similar articles
-
Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.Science. 2018 Nov 2;362(6414):598-602. doi: 10.1126/science.aaq0620. Science. 2018. PMID: 30385580 Free PMC article.
-
Mapping of a Novel H3-Specific Broadly Neutralizing Monoclonal Antibody Targeting the Hemagglutinin Globular Head Isolated from an Elite Influenza Virus-Immunized Donor Exhibiting Serological Breadth.J Virol. 2020 Feb 28;94(6):e01035-19. doi: 10.1128/JVI.01035-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31826999 Free PMC article.
-
A highly conserved neutralizing epitope on group 2 influenza A viruses.Science. 2011 Aug 12;333(6044):843-50. doi: 10.1126/science.1204839. Epub 2011 Jul 7. Science. 2011. PMID: 21737702 Free PMC article.
-
Implications of broadly neutralizing antibodies in the development of a universal influenza vaccine.Curr Opin Virol. 2016 Apr;17:110-115. doi: 10.1016/j.coviro.2016.03.002. Epub 2016 Mar 28. Curr Opin Virol. 2016. PMID: 27031684 Free PMC article. Review.
-
Influenza virus glycoprotein-reactive human monoclonal antibodies.Microbes Infect. 2020 Jul-Aug;22(6-7):263-271. doi: 10.1016/j.micinf.2020.06.003. Epub 2020 Jun 19. Microbes Infect. 2020. PMID: 32569735 Free PMC article. Review.
Cited by
-
Mechanistic dissection of antibody inhibition of influenza entry yields unexpected heterogeneity.Biophys J. 2023 Jun 6;122(11):1996-2006. doi: 10.1016/j.bpj.2022.10.026. Epub 2022 Oct 19. Biophys J. 2023. PMID: 36262043 Free PMC article.
-
A chimeric mRNA vaccine of S-RBD with HA conferring broad protection against influenza and COVID-19 variants.PLoS Pathog. 2024 Sep 20;20(9):e1012508. doi: 10.1371/journal.ppat.1012508. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39303003 Free PMC article.
-
Broad Reactivity Single Domain Antibodies against Influenza Virus and Their Applications to Vaccine Potency Testing and Immunotherapy.Biomolecules. 2021 Mar 10;11(3):407. doi: 10.3390/biom11030407. Biomolecules. 2021. PMID: 33802072 Free PMC article. Review.
-
Maturation of germinal center B cells after influenza virus vaccination in humans.J Exp Med. 2024 Aug 5;221(8):e20240668. doi: 10.1084/jem.20240668. Epub 2024 Jun 27. J Exp Med. 2024. PMID: 38935072 Free PMC article.
-
Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.PLoS Pathog. 2013;9(5):e1003342. doi: 10.1371/journal.ppat.1003342. Epub 2013 May 2. PLoS Pathog. 2013. PMID: 23658524 Free PMC article.
References
-
- WHO. Fact sheet 211: Influenza. 2009
-
- Lowen AC, Palese P. Influenza virus transmission: basic science and implications for the use of antiviral drugs during a pandemic. Infect Disord Drug Targets. 2007;7:318–328. - PubMed
-
- Wang TT, Palese P. Universal epitopes of influenza virus hemagglutinins? Nature Struct Biol. 2009;16:233–234. - PubMed
-
- Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

