MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame
- PMID: 22870278
- PMCID: PMC3411609
- DOI: 10.1371/journal.pone.0042034
MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame
Abstract
Background: MDM4, also called MDMX or HDMX in humans, is an important negative regulator of the p53 tumor suppressor. MDM4 is overexpressed in about 17% of all cancers and more frequently in some types, such as colon cancer or retinoblastoma. MDM4 is known to be post-translationally regulated by MDM2-mediated ubiquitination to decrease its protein levels in response to genotoxic stress, resulting in accumulation and activation of p53. At the transcriptional level, MDM4 gene regulation has been less clearly understood. We have reported that DNA damage triggers loss of MDM4 mRNA and a concurrent increase in p53 activity. These experiments attempt to determine a mechanism for down-regulation of MDM4 mRNA.
Methodology/principal findings: Here we report that MDM4 mRNA is a target of hsa-mir-34a (miR-34a). MDM4 mRNA contains a lengthy 3' untranslated region; however, we find that it is a miR-34a site within the open reading frame (ORF) of exon 11 that is responsible for the repression. Overexpression of miR-34a, but not a mutant miR-34a, is sufficient to decrease MDM4 mRNA levels to an extent identical to those of known miR-34a target genes. Likewise, MDM4 protein levels are decreased by miR-34a overexpression. Inhibition of endogenous miR-34a increased expression of miR-34a target genes and MDM4. A portion of MDM4 exon 11 containing this 8mer-A1 miR-34a site fused to a luciferase reporter gene is sufficient to confer responsiveness, being inhibited by additional expression of exogenous mir-34a and activated by inhibition of miR-34a.
Conclusions/significance: These data establish a mechanism for the observed DNA damage-induced negative regulation of MDM4 and potentially provide a novel means to manipulate MDM4 expression without introducing DNA damage.
Conflict of interest statement
Figures
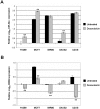

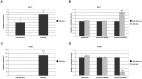

Similar articles
-
miR-661 downregulates both Mdm2 and Mdm4 to activate p53.Cell Death Differ. 2014 Feb;21(2):302-9. doi: 10.1038/cdd.2013.146. Epub 2013 Oct 18. Cell Death Differ. 2014. PMID: 24141721 Free PMC article.
-
Luteolin Inhibits Tumorigenesis and Induces Apoptosis of Non-Small Cell Lung Cancer Cells via Regulation of MicroRNA-34a-5p.Int J Mol Sci. 2018 Feb 2;19(2):447. doi: 10.3390/ijms19020447. Int J Mol Sci. 2018. PMID: 29393891 Free PMC article.
-
An illegitimate microRNA target site within the 3' UTR of MDM4 affects ovarian cancer progression and chemosensitivity.Cancer Res. 2010 Dec 1;70(23):9641-9. doi: 10.1158/0008-5472.CAN-10-0527. Epub 2010 Nov 16. Cancer Res. 2010. PMID: 21084273
-
Regulation of MDM4.Front Biosci (Landmark Ed). 2011 Jan 1;16(3):1144-56. doi: 10.2741/3780. Front Biosci (Landmark Ed). 2011. PMID: 21196223 Review.
-
MDM4: What do we know about the association between its polymorphisms and cancer?Med Oncol. 2022 Dec 24;40(1):61. doi: 10.1007/s12032-022-01929-z. Med Oncol. 2022. PMID: 36566308 Review.
Cited by
-
DNA methylation and miRNA-1296 act in concert to mediate spatiotemporal expression of KPNA7 during bovine oocyte and early embryonic development.BMC Dev Biol. 2019 Dec 2;19(1):23. doi: 10.1186/s12861-019-0204-x. BMC Dev Biol. 2019. PMID: 31787077 Free PMC article.
-
A feedback circuit of miR-34a/MDM4/p53 regulates apoptosis in chronic lymphocytic leukemia cells.Transl Cancer Res. 2020 Oct;9(10):6143-6153. doi: 10.21037/tcr-20-1710. Transl Cancer Res. 2020. PMID: 35117225 Free PMC article.
-
A comprehensive investigation using meta-analysis and bioinformatics on miR-34a-5p expression and its potential role in head and neck squamous cell carcinoma.Am J Transl Res. 2018 Aug 15;10(8):2246-2263. eCollection 2018. Am J Transl Res. 2018. PMID: 30210668 Free PMC article.
-
The p53/microRNA connection in gastrointestinal cancer.Clin Exp Gastroenterol. 2014 Sep 30;7:395-413. doi: 10.2147/CEG.S43738. eCollection 2014. Clin Exp Gastroenterol. 2014. PMID: 25328413 Free PMC article. Review.
-
MicroRNAs in B-cells: from normal differentiation to treatment of malignancies.Oncotarget. 2015 Jan 1;6(1):7-25. doi: 10.18632/oncotarget.3057. Oncotarget. 2015. PMID: 25622103 Free PMC article. Review.
References
-
- Mancini F, Moretti F (2009) Mitochondrial MDM4 (MDMX): an unpredicted role in the p53-mediated intrinsic apoptotic pathway. Cell Cycle 8: 3854–3859. - PubMed
-
- Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, et al. (2006) A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell 9: 273–285. - PubMed
-
- Rickman DS, Bobek MP, Misek DE, Kuick R, Blaivas M, et al. (2001) Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res 61: 6885–6891. - PubMed
-
- Riemenschneider MJ, Buschges R, Wolter M, Reifenberger J, Bostrom J, et al. (1999) Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res 59: 6091–6096. - PubMed
-
- Riemenschneider MJ, Knobbe CB, Reifenberger G (2003) Refined mapping of 1q32 amplicons in malignant gliomas confirms MDM4 as the main amplification target. Int J Cancer 104: 752–757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

