Stabilization of HIF-2α through redox regulation of mTORC2 activation and initiation of mRNA translation
- PMID: 22869144
- PMCID: PMC3696051
- DOI: 10.1038/onc.2012.333
Stabilization of HIF-2α through redox regulation of mTORC2 activation and initiation of mRNA translation
Abstract
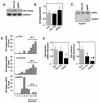
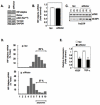
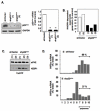
Hypoxia inducible factor-2α (HIF-2α) has a critical role in renal tumorigenesis. HIF-2α is stabilized in von Hippel-Lindau (VHL)-deficient renal cell carcinoma through mechanisms that require ongoing mRNA translation. Mammalian target of rapamycin (mTOR) functions in two distinct complexes: Raptor-associated mTORC1 and Rictor-associated mTORC2. Rictor-associated mTORC2 complex has been linked to maintaining HIF-2α protein in the absence of VHL; however, the mechanisms remain to be elucidated. Although Raptor-associated mTORC1 is a known key upstream regulator of mRNA translation, initiation and elongation, the role of mTORC2 in regulating mRNA translation is not clear. Complex assembly of the mRNA cap protein, eukaryotic translation initiation factor 4 (eIF4)E, with activators (eIF4 gamma (eIF4G)) and inhibitors (eIF4E-binding protein 1 (4E-BP1)) are rate-limiting determinants of mRNA translation. Our laboratory has previously demonstrated that reactive oxygen species, mediated by p22(phox)-based Nox oxidases, are enhanced in VHL-deficient cells and have a role in the activation of Akt on S473, a site phosphorylated by the mTORC2 complex. In this study, we examined the role of Rictor-dependent regulation of HIF-2α through eIF4E-dependent mRNA translation and examined the effects of p22(phox)-based Nox oxidases on TORC2 regulation. We demonstrate for the first time that mTORC2 complex stability and activation is redox sensitive, and further defined a novel role for p22(phox)-based Nox oxidases in eIF4E-dependent mRNA translation through mTORC2. Furthermore, we provide the first evidence that silencing of p22(phox) reduces HIF-2α-dependent gene targeting in vitro and tumor formation in vivo. The clinical relevance of these studies is demonstrated.
Figures
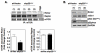


Similar articles
-
The NADPH oxidase subunit p22phox inhibits the function of the tumor suppressor protein tuberin.Am J Pathol. 2010 May;176(5):2447-55. doi: 10.2353/ajpath.2010.090606. Epub 2010 Mar 19. Am J Pathol. 2010. PMID: 20304964 Free PMC article.
-
Chemotherapy-mediated p53-dependent DNA damage response in clear cell renal cell carcinoma: role of the mTORC1/2 and hypoxia-inducible factor pathways.Cell Death Dis. 2013 Oct 17;4(10):e865. doi: 10.1038/cddis.2013.395. Cell Death Dis. 2013. PMID: 24136229 Free PMC article.
-
Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function.Oncogene. 2000 Nov 16;19(48):5435-43. doi: 10.1038/sj.onc.1203938. Oncogene. 2000. PMID: 11114720
-
VHL and HIF signalling in renal cell carcinogenesis.J Pathol. 2010 Jun;221(2):125-38. doi: 10.1002/path.2689. J Pathol. 2010. PMID: 20225241 Review.
-
Recent progress in the development of hypoxia-inducible factor 2α (HIF-2α) modulators: Inhibitors, agonists, and degraders (2009-2024).Eur J Med Chem. 2024 Sep 5;275:116645. doi: 10.1016/j.ejmech.2024.116645. Epub 2024 Jul 1. Eur J Med Chem. 2024. PMID: 38959730 Review.
Cited by
-
Complex interplay between autophagy and oxidative stress in the development of pulmonary disease.Redox Biol. 2020 Sep;36:101679. doi: 10.1016/j.redox.2020.101679. Epub 2020 Aug 11. Redox Biol. 2020. PMID: 32818797 Free PMC article. Review.
-
Nitration of MnSOD in the Carotid Body and Adrenal Gland Induced by Chronic Intermittent Hypoxia.J Histochem Cytochem. 2018 Oct;66(10):753-765. doi: 10.1369/0022155418776229. Epub 2018 May 18. J Histochem Cytochem. 2018. PMID: 29775122 Free PMC article.
-
The antioxidant paradox: what are antioxidants and how should they be used in a therapeutic context for cancer.Future Med Chem. 2014;6(12):1413-22. doi: 10.4155/fmc.14.86. Future Med Chem. 2014. PMID: 25329197 Free PMC article. Review.
-
NOX4 functions as a mitochondrial energetic sensor coupling cancer metabolic reprogramming to drug resistance.Nat Commun. 2017 Oct 19;8(1):997. doi: 10.1038/s41467-017-01106-1. Nat Commun. 2017. PMID: 29051480 Free PMC article.
-
Icariside II, a natural mTOR inhibitor, disrupts aberrant energy homeostasis via suppressing mTORC1-4E-BP1 axis in sarcoma cells.Oncotarget. 2016 May 10;7(19):27819-37. doi: 10.18632/oncotarget.8538. Oncotarget. 2016. PMID: 27056897 Free PMC article.
References
-
- Kaelin WG., Jr. The von Hippel-Lindau tumor suppressor protein and clear cell renal carcinoma. Clin Cancer Res. 2007 Jan 15;13(2 Pt 2):680s–684s. - PubMed
-
- Baldewijns MM, van Vlodrop IJ, Vermeulen PB, Soetekouw PM, van Engeland M, de Bruïne AP. VHL and HIF signalling in renal cell carcinogenesis. J Pathol. 2010 Jun;221(2):125–38. Review. - PubMed
-
- Block K, Gorin Y, Hoover P, Williams P, Chelmicki T, Clark RA, et al. NAD(P)H oxidases regulate HIF-2alpha protein expression. J Biol Chem. 2007 Mar 16;282(11):8019–26. - PubMed
-
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990 Jun 7;345(6275):544–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

