Structure and function of biotin-dependent carboxylases
- PMID: 22869039
- PMCID: PMC3508090
- DOI: 10.1007/s00018-012-1096-0
Structure and function of biotin-dependent carboxylases
Abstract
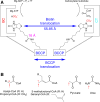
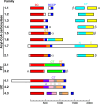
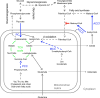
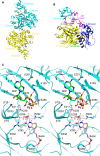
Biotin-dependent carboxylases include acetyl-CoA carboxylase (ACC), propionyl-CoA carboxylase (PCC), 3-methylcrotonyl-CoA carboxylase (MCC), geranyl-CoA carboxylase, pyruvate carboxylase (PC), and urea carboxylase (UC). They contain biotin carboxylase (BC), carboxyltransferase (CT), and biotin-carboxyl carrier protein components. These enzymes are widely distributed in nature and have important functions in fatty acid metabolism, amino acid metabolism, carbohydrate metabolism, polyketide biosynthesis, urea utilization, and other cellular processes. ACCs are also attractive targets for drug discovery against type 2 diabetes, obesity, cancer, microbial infections, and other diseases, and the plastid ACC of grasses is the target of action of three classes of commercial herbicides. Deficiencies in the activities of PCC, MCC, or PC are linked to serious diseases in humans. Our understanding of these enzymes has been greatly enhanced over the past few years by the crystal structures of the holoenzymes of PCC, MCC, PC, and UC. The structures reveal unanticipated features in the architectures of the holoenzymes, including the presence of previously unrecognized domains, and provide a molecular basis for understanding their catalytic mechanism as well as the large collection of disease-causing mutations in PCC, MCC, and PC. This review will summarize the recent advances in our knowledge on the structure and function of these important metabolic enzymes.
Figures





Similar articles
-
Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase.Nature. 2010 Aug 19;466(7309):1001-5. doi: 10.1038/nature09302. Nature. 2010. PMID: 20725044 Free PMC article.
-
Striking Diversity in Holoenzyme Architecture and Extensive Conformational Variability in Biotin-Dependent Carboxylases.Adv Protein Chem Struct Biol. 2017;109:161-194. doi: 10.1016/bs.apcsb.2017.04.006. Epub 2017 May 23. Adv Protein Chem Struct Biol. 2017. PMID: 28683917 Review.
-
Chloroflexus aurantiacus acetyl-CoA carboxylase evolves fused biotin carboxylase and biotin carboxyl carrier protein to complete carboxylation activity.mBio. 2024 May 8;15(5):e0341423. doi: 10.1128/mbio.03414-23. Epub 2024 Apr 4. mBio. 2024. PMID: 38572988 Free PMC article.
-
Purification and characterization of mitochondrial biotin-dependent carboxylases from native tissues.Methods Enzymol. 2024;708:1-30. doi: 10.1016/bs.mie.2024.10.010. Epub 2024 Oct 28. Methods Enzymol. 2024. PMID: 39572135
-
The biotin enzyme family: conserved structural motifs and domain rearrangements.Curr Protein Pept Sci. 2003 Jun;4(3):217-29. doi: 10.2174/1389203033487199. Curr Protein Pept Sci. 2003. PMID: 12769720 Review.
Cited by
-
Crystal structures of malonyl-coenzyme A decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.Structure. 2013 Jul 2;21(7):1182-92. doi: 10.1016/j.str.2013.05.001. Epub 2013 Jun 20. Structure. 2013. PMID: 23791943 Free PMC article.
-
Characterization of the mycobacterial acyl-CoA carboxylase holo complexes reveals their functional expansion into amino acid catabolism.PLoS Pathog. 2015 Feb 19;11(2):e1004623. doi: 10.1371/journal.ppat.1004623. eCollection 2015 Feb. PLoS Pathog. 2015. PMID: 25695631 Free PMC article.
-
A substrate-induced biotin binding pocket in the carboxyltransferase domain of pyruvate carboxylase.J Biol Chem. 2013 Jul 5;288(27):19915-25. doi: 10.1074/jbc.M113.477828. Epub 2013 May 22. J Biol Chem. 2013. PMID: 23698000 Free PMC article.
-
Enhancing Multistep Reactions: Biomimetic Design of Substrate Channeling Using P22 Virus-Like Particles.Adv Sci (Weinh). 2023 May;10(13):e2206906. doi: 10.1002/advs.202206906. Epub 2023 Feb 23. Adv Sci (Weinh). 2023. PMID: 36815387 Free PMC article.
-
Testosterone Degradative Pathway of Novosphingobium tardaugens.Genes (Basel). 2019 Oct 31;10(11):871. doi: 10.3390/genes10110871. Genes (Basel). 2019. PMID: 31683600 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

