RCrane: semi-automated RNA model building
- PMID: 22868764
- PMCID: PMC3413212
- DOI: 10.1107/S0907444912018549
RCrane: semi-automated RNA model building
Abstract
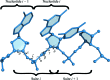
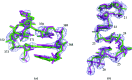
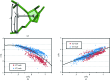
RNA crystals typically diffract to much lower resolutions than protein crystals. This low-resolution diffraction results in unclear density maps, which cause considerable difficulties during the model-building process. These difficulties are exacerbated by the lack of computational tools for RNA modeling. Here, RCrane, a tool for the partially automated building of RNA into electron-density maps of low or intermediate resolution, is presented. This tool works within Coot, a common program for macromolecular model building. RCrane helps crystallographers to place phosphates and bases into electron density and then automatically predicts and builds the detailed all-atom structure of the traced nucleotides. RCrane then allows the crystallographer to review the newly built structure and select alternative backbone conformations where desired. This tool can also be used to automatically correct the backbone structure of previously built nucleotides. These automated corrections can fix incorrect sugar puckers, steric clashes and other structural problems.
Figures

Similar articles
-
Semiautomated model building for RNA crystallography using a directed rotameric approach.Proc Natl Acad Sci U S A. 2010 May 4;107(18):8177-82. doi: 10.1073/pnas.0911888107. Epub 2010 Apr 19. Proc Natl Acad Sci U S A. 2010. PMID: 20404211 Free PMC article.
-
Automated crystallographic ligand building using the medial axis transform of an electron-density isosurface.Acta Crystallogr D Biol Crystallogr. 2005 Oct;61(Pt 10):1354-63. doi: 10.1107/S0907444905023152. Epub 2005 Sep 28. Acta Crystallogr D Biol Crystallogr. 2005. PMID: 16204887
-
Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7.Nat Protoc. 2008;3(7):1171-9. doi: 10.1038/nprot.2008.91. Nat Protoc. 2008. PMID: 18600222 Free PMC article.
-
Macromolecular modeling with rosetta.Annu Rev Biochem. 2008;77:363-82. doi: 10.1146/annurev.biochem.77.062906.171838. Annu Rev Biochem. 2008. PMID: 18410248 Review.
-
Interactive model building in neutron macromolecular crystallography.Methods Enzymol. 2020;634:201-224. doi: 10.1016/bs.mie.2019.11.017. Epub 2019 Dec 20. Methods Enzymol. 2020. PMID: 32093833 Review.
Cited by
-
Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions.Acta Crystallogr D Biol Crystallogr. 2015 Jan 1;71(Pt 1):136-53. doi: 10.1107/S1399004714021683. Epub 2015 Jan 1. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25615868 Free PMC article.
-
Crystal structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) frameshifting pseudoknot.RNA. 2022 Feb;28(2):239-249. doi: 10.1261/rna.078825.121. Epub 2021 Nov 29. RNA. 2022. PMID: 34845084 Free PMC article.
-
Visualizing the ai5γ group IIB intron.Nucleic Acids Res. 2014 Feb;42(3):1947-58. doi: 10.1093/nar/gkt1051. Epub 2013 Nov 6. Nucleic Acids Res. 2014. PMID: 24203709 Free PMC article.
-
Crystal structure of a eukaryotic group II intron lariat.Nature. 2014 Oct 9;514(7521):193-7. doi: 10.1038/nature13790. Epub 2014 Sep 24. Nature. 2014. PMID: 25252982 Free PMC article.
-
Automated nucleic acid chain tracing in real time.IUCrJ. 2014 Sep 23;1(Pt 6):387-92. doi: 10.1107/S2052252514019290. eCollection 2014 Nov 1. IUCrJ. 2014. PMID: 25485119 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

