Spatio-temporal regulation of ADAR editing during development in porcine neural tissues
- PMID: 22858680
- PMCID: PMC3551860
- DOI: 10.4161/rna.21082
Spatio-temporal regulation of ADAR editing during development in porcine neural tissues
Abstract
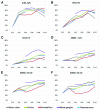
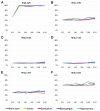
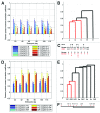
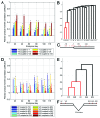
Editing by ADAR enzymes is essential for mammalian life. Still, knowledge of the spatio-temporal editing patterns in mammals is limited. By use of 454 amplicon sequencing we examined the editing status of 12 regionally extracted mRNAs from porcine developing brain encompassing a total of 64 putative ADAR editing sites. In total 24 brain tissues, dissected from up to five regions from embryonic gestation day 23, 42, 60, 80, 100 and 115, were examined for editing. Generally, editing increased during embryonic development concomitantly with an increase in ADAR2 mRNA level. Notably, the Gria2 (GluR-B) Q/R site, reported to be ~100% edited in previous studies, is only 54% edited at embryonic day 23. Transcripts with multiple editing sites in close proximity to each other exhibit coupled editing and an extraordinary incidence of long-range coupling of editing events more than 32 kb apart is observed for the kainate glutamate receptor 2 transcript, Grik2. Our study reveals complex spatio-temporal ADAR editing patterns of coordinated editing events that may play important roles in the development of the mammalian brain.
Figures

Similar articles
-
Adenosine Deaminase That Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-mRNA Inhibits RNA Editing in Glioblastoma.J Biol Chem. 2017 Mar 10;292(10):4326-4335. doi: 10.1074/jbc.M117.779868. Epub 2017 Feb 6. J Biol Chem. 2017. PMID: 28167531 Free PMC article.
-
A comparative analysis of ADAR mutant mice reveals site-specific regulation of RNA editing.RNA. 2020 Apr;26(4):454-469. doi: 10.1261/rna.072728.119. Epub 2020 Jan 15. RNA. 2020. PMID: 31941663 Free PMC article.
-
Developmental changes in expression and self-editing of adenosine deaminase type 2 pre-mRNA and mRNA in rat brain and cultured cortical neurons.Neurosci Res. 2008 Aug;61(4):398-403. doi: 10.1016/j.neures.2008.04.007. Epub 2008 Apr 26. Neurosci Res. 2008. PMID: 18534702
-
ADAR RNA editing in innate immune response phasing, in circadian clocks and in sleep.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):356-369. doi: 10.1016/j.bbagrm.2018.10.011. Epub 2018 Oct 31. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30391332 Review.
-
Regulation and functions of ADAR in drosophila.Curr Top Microbiol Immunol. 2012;353:221-36. doi: 10.1007/82_2011_152. Curr Top Microbiol Immunol. 2012. PMID: 21761288 Review.
Cited by
-
Roles of miR-432 and circ_0000418 in mediating the anti-depressant action of ADAR1.Neurobiol Stress. 2021 Sep 11;15:100396. doi: 10.1016/j.ynstr.2021.100396. eCollection 2021 Nov. Neurobiol Stress. 2021. PMID: 34568523 Free PMC article.
-
RNA editing differently affects protein-coding genes in D. melanogaster and H. sapiens.Sci Rep. 2015 Jul 14;5:11550. doi: 10.1038/srep11550. Sci Rep. 2015. PMID: 26169954 Free PMC article.
-
Genome-wide identification of RNA editing in seven porcine tissues by matched DNA and RNA high-throughput sequencing.J Anim Sci Biotechnol. 2019 Mar 13;10:24. doi: 10.1186/s40104-019-0326-9. eCollection 2019. J Anim Sci Biotechnol. 2019. PMID: 30911384 Free PMC article.
-
Developmental atlas of the RNA editome in Sus scrofa skeletal muscle.DNA Res. 2019 Jun 1;26(3):261-272. doi: 10.1093/dnares/dsz006. DNA Res. 2019. PMID: 31231762 Free PMC article.
-
Adenosine-to-inosine RNA editing in neurological development and disease.RNA Biol. 2021 Jul;18(7):999-1013. doi: 10.1080/15476286.2020.1867797. Epub 2021 Jan 6. RNA Biol. 2021. PMID: 33393416 Free PMC article. Review.
References
-
- George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A. 1999;96:4621–6. doi: 10.1073/pnas.96.8.4621. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
