Bullied no more: when and how DNA shoves proteins around
- PMID: 22850561
- PMCID: PMC4866820
- DOI: 10.1017/S0033583512000054
Bullied no more: when and how DNA shoves proteins around
Abstract
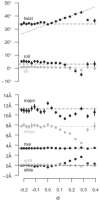
The predominant protein-centric perspective in protein-DNA-binding studies assumes that the protein drives the interaction. Research focuses on protein structural motifs, electrostatic surfaces and contact potentials, while DNA is often ignored as a passive polymer to be manipulated. Recent studies of DNA topology, the supercoiling, knotting, and linking of the helices, have shown that DNA has the capability to be an active participant in its transactions. DNA topology-induced structural and geometric changes can drive, or at least strongly influence, the interactions between protein and DNA. Deformations of the B-form structure arise from both the considerable elastic energy arising from supercoiling and from the electrostatic energy. Here, we discuss how these energies are harnessed for topology-driven, sequence-specific deformations that can allow DNA to direct its own metabolism.
Figures


Similar articles
-
Interference between Triplex and Protein Binding to Distal Sites on Supercoiled DNA.Biophys J. 2017 Feb 7;112(3):523-531. doi: 10.1016/j.bpj.2016.12.034. Epub 2017 Jan 17. Biophys J. 2017. PMID: 28108011 Free PMC article.
-
Varying levels of positive and negative supercoiling differently affect the efficiency with which topoisomerase II catenates and decatenates DNA.J Mol Biol. 2001 Jan 19;305(3):441-50. doi: 10.1006/jmbi.2000.4307. J Mol Biol. 2001. PMID: 11152602
-
Cellular strategies for regulating DNA supercoiling: a single-molecule perspective.Cell. 2010 Aug 20;142(4):519-30. doi: 10.1016/j.cell.2010.08.001. Cell. 2010. PMID: 20723754 Free PMC article. Review.
-
Topological aspects of DNA function and protein folding.Biochem Soc Trans. 2013 Apr;41(2):491-3. doi: 10.1042/BST20130006. Biochem Soc Trans. 2013. PMID: 23514141
-
Protein tracking-induced supercoiling of DNA: a tool to regulate DNA transactions in vivo?Bioessays. 1994 Feb;16(2):91-9. doi: 10.1002/bies.950160205. Bioessays. 1994. PMID: 8147849 Review.
Cited by
-
Protein/DNA interactions in complex DNA topologies: expect the unexpected.Biophys Rev. 2016;8(3):233-243. doi: 10.1007/s12551-016-0208-8. Epub 2016 Aug 8. Biophys Rev. 2016. PMID: 27738452 Free PMC article. Review.
-
Searching target sites on DNA by proteins: Role of DNA dynamics under confinement.Nucleic Acids Res. 2015 Oct 30;43(19):9176-86. doi: 10.1093/nar/gkv931. Epub 2015 Sep 22. Nucleic Acids Res. 2015. PMID: 26400158 Free PMC article.
-
DNA supercoiling-induced shapes alter minicircle hydrodynamic properties.Nucleic Acids Res. 2023 May 8;51(8):4027-4042. doi: 10.1093/nar/gkad183. Nucleic Acids Res. 2023. PMID: 36971110 Free PMC article.
-
The Dynamic Interplay Between DNA Topoisomerases and DNA Topology.Biophys Rev. 2016 Sep;8(3):221-231. doi: 10.1007/s12551-016-0206-x. Epub 2016 Jul 2. Biophys Rev. 2016. PMID: 27942270 Free PMC article.
-
Chromosomal organization of transcription: in a nutshell.Curr Genet. 2018 Jun;64(3):555-565. doi: 10.1007/s00294-017-0785-5. Epub 2017 Nov 28. Curr Genet. 2018. PMID: 29184972 Review.
References
-
- Aggarwal A, Rodgers DW, Drottar M, Ptashne M, Harrison SC. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242:899–907. - PubMed
-
- Anderson P, Bauer W. Supercoiling in closed circular DNA: dependence upon ion type and concentration. Biochemistry. 1978;17:594–601. - PubMed
-
- Ansari AZ, Chael ML, O'Halloran TV. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-Mer. Nature. 1992;355:87–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
