Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis
- PMID: 22842725
- PMCID: PMC3414646
- DOI: 10.1038/nsmb.2347
Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis
Abstract
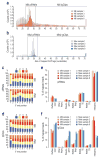
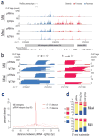
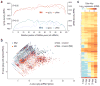
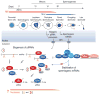
Germ cells implement elaborate mechanisms to protect their genetic material and to regulate gene expression during differentiation. Piwi proteins bind Piwi-interacting RNAs (piRNAs), small germline RNAs whose biogenesis and functions are still largely elusive. We used high-throughput sequencing after cross-linking and immunoprecipitation (HITS-CLIP) coupled with RNA-sequencing (RNA-seq) to characterize the genome-wide target RNA repertoire of Mili (Piwil2) and Miwi (Piwil1), two Piwi proteins expressed in mouse postnatal testis. We report the in vivo pathway of primary piRNA biogenesis and implicate distinct nucleolytic activities that process Piwi-bound precursor transcripts. Our studies indicate that pachytene piRNAs are the end products of RNA processing. HITS-CLIP demonstrated that Miwi binds spermiogenic mRNAs directly, without using piRNAs as guides, and independent biochemical analyses of testis mRNA ribonucleoproteins (mRNPs) established that Miwi functions in the formation of mRNP complexes that stabilize mRNAs essential for spermiogenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Mitochondrial membrane-based initial separation of MIWI and MILI functions during pachytene piRNA biogenesis.Nucleic Acids Res. 2019 Mar 18;47(5):2594-2608. doi: 10.1093/nar/gky1281. Nucleic Acids Res. 2019. PMID: 30590800 Free PMC article.
-
Identification of MIWI-associated Poly(A) RNAs by immunoprecipitation with an anti-MIWI monoclonal antibody.Biosci Trends. 2012 Oct;6(5):248-61. doi: 10.5582/bst.2012.v6.5.248. Biosci Trends. 2012. PMID: 23229118
-
MIWI N-terminal arginines orchestrate generation of functional pachytene piRNAs and spermiogenesis.Nucleic Acids Res. 2024 Jun 24;52(11):6558-6570. doi: 10.1093/nar/gkae193. Nucleic Acids Res. 2024. PMID: 38520410 Free PMC article.
-
The regulatory functions of piRNA/PIWI in spermatogenesis.Yi Chuan. 2017 Aug 20;39(8):683-691. doi: 10.16288/j.yczz.17-245. Yi Chuan. 2017. PMID: 28903896 Review.
-
Noncanonical functions of PIWIL1/piRNAs in animal male germ cells and human diseases†.Biol Reprod. 2022 Jul 25;107(1):101-108. doi: 10.1093/biolre/ioac073. Biol Reprod. 2022. PMID: 35403682 Review.
Cited by
-
FUS regulates genes coding for RNA-binding proteins in neurons by binding to their highly conserved introns.RNA. 2013 Apr;19(4):498-509. doi: 10.1261/rna.037804.112. Epub 2013 Feb 6. RNA. 2013. PMID: 23389473 Free PMC article.
-
RAN-binding protein 9 is involved in alternative splicing and is critical for male germ cell development and male fertility.PLoS Genet. 2014 Dec 4;10(12):e1004825. doi: 10.1371/journal.pgen.1004825. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25474150 Free PMC article.
-
Piwi Proteins and piRNAs step onto the systems biology stage.Adv Exp Med Biol. 2014;825:159-97. doi: 10.1007/978-1-4939-1221-6_5. Adv Exp Med Biol. 2014. PMID: 25201106 Free PMC article.
-
Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes.Nucleic Acids Res. 2017 May 19;45(9):5387-5398. doi: 10.1093/nar/gkx027. Nucleic Acids Res. 2017. PMID: 28115634 Free PMC article.
-
The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline.Elife. 2017 Oct 31;6:e26116. doi: 10.7554/eLife.26116. Elife. 2017. PMID: 29087293 Free PMC article.
References
-
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–30. - PubMed
-
- Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–49. - PubMed
-
- Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. - PubMed
-
- Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

