Genomic and serological detection of bat coronavirus from bats in the Philippines
- PMID: 22833101
- PMCID: PMC7086765
- DOI: 10.1007/s00705-012-1410-z
Genomic and serological detection of bat coronavirus from bats in the Philippines
Abstract
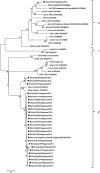
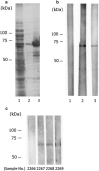
Bat coronavirus (BtCoV) is assumed to be a progenitor of severe acute respiratory syndrome (SARS)-related coronaviruses. To explore the distribution of BtCoVs in the Philippines, we collected 179 bats and detected viral RNA from intestinal or fecal samples by RT-PCR. The overall prevalence of BtCoVs among bats was 29.6 %. Phylogenetic analysis of the partial RNA-dependent RNA polymerase gene suggested that one of the detected BtCoVs was a novel alphacoronavirus, while the others belonged to the genus Betacoronavirus. Western blotting revealed that 66.5 % of bat sera had antibodies to BtCoV. These surveys suggested the endemic presence of BtCoVs in the Philippines.
Figures
Similar articles
-
Detection of bat coronaviruses from Miniopterus fuliginosus in Japan.Virus Genes. 2012 Feb;44(1):40-4. doi: 10.1007/s11262-011-0661-1. Epub 2011 Aug 30. Virus Genes. 2012. PMID: 21877208 Free PMC article.
-
Bat coronaviruses and experimental infection of bats, the Philippines.Emerg Infect Dis. 2010 Aug;16(8):1217-23. doi: 10.3201/eid1608.100208. Emerg Infect Dis. 2010. PMID: 20678314 Free PMC article.
-
Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft.Virol Sin. 2016 Feb;31(1):31-40. doi: 10.1007/s12250-016-3713-9. Epub 2016 Feb 18. Virol Sin. 2016. PMID: 26920708 Free PMC article.
-
Coronaviruses in Bats: A Review for the Americas.Viruses. 2021 Jun 25;13(7):1226. doi: 10.3390/v13071226. Viruses. 2021. PMID: 34201926 Free PMC article. Review.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
Cited by
-
Global Epidemiology of Bat Coronaviruses.Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. Viruses. 2019. PMID: 30791586 Free PMC article. Review.
-
Longitudinal study of age-specific pattern of coronavirus infection in Lyle's flying fox (Pteropus lylei) in Thailand.Virol J. 2018 Feb 20;15(1):38. doi: 10.1186/s12985-018-0950-6. Virol J. 2018. PMID: 29463282 Free PMC article.
-
Longitudinal monitoring in Cambodia suggests higher circulation of alpha and betacoronaviruses in juvenile and immature bats of three species.Sci Rep. 2021 Dec 17;11(1):24145. doi: 10.1038/s41598-021-03169-z. Sci Rep. 2021. PMID: 34921180 Free PMC article.
-
Overview of Bat and Wildlife Coronavirus Surveillance in Africa: A Framework for Global Investigations.Viruses. 2021 May 18;13(5):936. doi: 10.3390/v13050936. Viruses. 2021. PMID: 34070175 Free PMC article. Review.
-
Detection of coronavirus genomes in Moluccan naked-backed fruit bats in Indonesia.Arch Virol. 2015 Apr;160(4):1113-8. doi: 10.1007/s00705-015-2342-1. Epub 2015 Feb 4. Arch Virol. 2015. PMID: 25643817 Free PMC article.
References
-
- Cc H, Hk V, Ds R, Tony S, Paul C. Bats prove to be rich reservoirs for emerging viruses. Microbe. 2008;3:521–528.
-
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguière AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
-
- WHO (2003) Cumulative number of reported probable cases of Severe Acute Respiratory Syndrome (SARS). http://www.who.int/csr/sars/country/2003_05_20/en/. Accessed 27 November 2011
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

