Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice
- PMID: 22826298
- PMCID: PMC3409501
- DOI: 10.1084/jem.20120504
Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice
Abstract
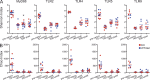
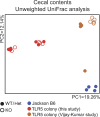
The intestinal microbiota contributes to the development of the immune system, and conversely, the immune system influences the composition of the microbiota. Toll-like receptors (TLRs) in the gut recognize bacterial ligands. Although TLR signaling represents a major arm of the innate immune system, the extent to which TLRs influence the composition of the intestinal microbiota remains unclear. We performed deep 16S ribosomal RNA sequencing to characterize the complex bacterial populations inhabiting the ileum and cecum of TLR- and MyD88-deficient mice. The microbiota of MyD88- and TLR-deficient mouse colonies differed markedly, with each colony harboring distinct and distinguishable bacterial populations in the small and large intestine. Comparison of MyD88-, TLR2-, TLR4-, TLR5-, and TLR9-deficient mice and their respective wild-type (WT) littermates demonstrated that the impact of TLR deficiency on the composition of the intestinal microbiota is minimal under homeostatic conditions and after recovery from antibiotic treatment. Thus, differences between TLR-deficient mouse colonies reflected long-term divergence of the microbiota after extended husbandry in isolation from each other. Long-term breeding of isolated mouse colonies results in changes of the intestinal microbiota that are communicated to offspring by maternal transmission, which account for marked compositional differences between WT and mutant mouse strains.
Figures


Similar articles
-
Pathogenic and protective roles of MyD88 in leukocytes and epithelial cells in mouse models of inflammatory bowel disease.Gastroenterology. 2010 Aug;139(2):519-29, 529.e1-2. doi: 10.1053/j.gastro.2010.04.045. Epub 2010 Apr 28. Gastroenterology. 2010. PMID: 20433840 Free PMC article.
-
Nucleic Acid-Sensing Toll-Like Receptors Play a Dominant Role in Innate Immune Recognition of Pneumococci.mBio. 2020 Mar 24;11(2):e00415-20. doi: 10.1128/mBio.00415-20. mBio. 2020. PMID: 32209688 Free PMC article.
-
Interleukin-1β (IL-1β) promotes susceptibility of Toll-like receptor 5 (TLR5) deficient mice to colitis.Gut. 2012 Mar;61(3):373-84. doi: 10.1136/gut.2011.240556. Epub 2011 Jun 5. Gut. 2012. PMID: 21646247
-
Dendritic cell specific targeting of MyD88 signalling pathways in vivo.Eur J Immunol. 2015 Jan;45(1):32-9. doi: 10.1002/eji.201444747. Epub 2014 Dec 16. Eur J Immunol. 2015. PMID: 25403892 Review.
-
Toll-like receptors and their signaling mechanism in innate immunity.Acta Odontol Scand. 2001 Jun;59(3):124-30. doi: 10.1080/000163501750266701. Acta Odontol Scand. 2001. PMID: 11501880 Review.
Cited by
-
Securing the border: lymphotoxin, IL-23 and IL-22 keep out the bad guys and 'fatten' the homeland.Nat Immunol. 2012 Oct;13(10):940-1. doi: 10.1038/ni.2425. Nat Immunol. 2012. PMID: 22990898 No abstract available.
-
Diet induced obesity is independent of metabolic endotoxemia and TLR4 signalling, but markedly increases hypothalamic expression of the acute phase protein, SerpinA3N.Sci Rep. 2018 Oct 23;8(1):15648. doi: 10.1038/s41598-018-33928-4. Sci Rep. 2018. PMID: 30353127 Free PMC article.
-
A computational analysis of dynamic, multi-organ inflammatory crosstalk induced by endotoxin in mice.PLoS Comput Biol. 2018 Nov 6;14(11):e1006582. doi: 10.1371/journal.pcbi.1006582. eCollection 2018 Nov. PLoS Comput Biol. 2018. PMID: 30399158 Free PMC article.
-
Toll-like receptors 2 and 4 modulate autonomic control of heart rate and energy metabolism.Brain Behav Immun. 2014 Feb;36:90-100. doi: 10.1016/j.bbi.2013.10.013. Epub 2013 Oct 18. Brain Behav Immun. 2014. PMID: 24145051 Free PMC article.
-
Interaction between microbiota and immunity in health and disease.Cell Res. 2020 Jun;30(6):492-506. doi: 10.1038/s41422-020-0332-7. Epub 2020 May 20. Cell Res. 2020. PMID: 32433595 Free PMC article. Review.
References
-
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B. 57:289–300
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

