DNA and chromatin modification networks distinguish stem cell pluripotent ground states
- PMID: 22822199
- PMCID: PMC3494154
- DOI: 10.1074/mcp.M111.011114
DNA and chromatin modification networks distinguish stem cell pluripotent ground states
Abstract
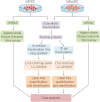
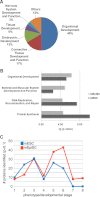
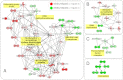
Pluripotent stem cells are capable of differentiating into all cell types of the body and therefore hold tremendous promise for regenerative medicine. Despite their widespread use in laboratories across the world, a detailed understanding of the molecular mechanisms that regulate the pluripotent state is currently lacking. Mouse embryonic (mESC) and epiblast (mEpiSC) stem cells are two closely related classes of pluripotent stem cells, derived from distinct embryonic tissues. Although both mESC and mEpiSC are pluripotent, these cell types show important differences in their properties suggesting distinct pluripotent ground states. To understand the molecular basis of pluripotency, we analyzed the nuclear proteomes of mESCs and mEpiSCs to identify protein networks that regulate their respective pluripotent states. Our study used label-free LC-MS/MS to identify and quantify 1597 proteins in embryonic and epiblast stem cell nuclei. Immunoblotting of a selected protein subset was used to confirm that key components of chromatin regulatory networks are differentially expressed in mESCs and mEpiSCs. Specifically, we identify differential expression of DNA methylation, ATP-dependent chromatin remodeling and nucleosome remodeling networks in mESC and mEpiSC nuclei. This study is the first comparative study of protein networks in cells representing the two distinct, pluripotent states, and points to the importance of DNA and chromatin modification processes in regulating pluripotency. In addition, by integrating our data with existing pluripotency networks, we provide detailed maps of protein networks that regulate pluripotency that will further both the fundamental understanding of pluripotency as well as efforts to reliably control the differentiation of these cells into functional cell fates.
Figures

Similar articles
-
The many faces of Pluripotency: in vitro adaptations of a continuum of in vivo states.BMC Dev Biol. 2017 Jun 13;17(1):7. doi: 10.1186/s12861-017-0150-4. BMC Dev Biol. 2017. PMID: 28610558 Free PMC article. Review.
-
iTRAQ proteome analysis reflects a progressed differentiation state of epiblast derived versus inner cell mass derived murine embryonic stem cells.J Proteomics. 2013 Sep 2;90:38-51. doi: 10.1016/j.jprot.2013.03.015. Epub 2013 Apr 18. J Proteomics. 2013. PMID: 23603003
-
Differential localization patterns of pyruvate kinase isoforms in murine naïve, formative, and primed pluripotent states.Exp Cell Res. 2021 Aug 15;405(2):112714. doi: 10.1016/j.yexcr.2021.112714. Epub 2021 Jun 26. Exp Cell Res. 2021. PMID: 34181938
-
E-cadherin promotes incorporation of mouse epiblast stem cells into normal development.PLoS One. 2012;7(9):e45220. doi: 10.1371/journal.pone.0045220. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028858 Free PMC article.
-
The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states.Curr Opin Genet Dev. 2013 Oct;23(5):504-11. doi: 10.1016/j.gde.2013.06.003. Epub 2013 Aug 7. Curr Opin Genet Dev. 2013. PMID: 23932125 Free PMC article. Review.
Cited by
-
Cdx2 efficiently induces trophoblast stem-like cells in naïve, but not primed, pluripotent stem cells.Stem Cells Dev. 2015 Jun 1;24(11):1352-65. doi: 10.1089/scd.2014.0395. Epub 2015 Mar 10. Stem Cells Dev. 2015. PMID: 25625326 Free PMC article.
-
RISC-mediated control of selected chromatin regulators stabilizes ground state pluripotency of mouse embryonic stem cells.Genome Biol. 2016 May 6;17(1):94. doi: 10.1186/s13059-016-0952-x. Genome Biol. 2016. PMID: 27154007 Free PMC article.
-
Epiblast-like stem cells established by Wnt/β-catenin signaling manifest distinct features of formative pluripotency and germline competence.Cell Rep. 2023 Jan 31;42(1):112021. doi: 10.1016/j.celrep.2023.112021. Epub 2023 Jan 23. Cell Rep. 2023. PMID: 36848234 Free PMC article.
-
The many faces of Pluripotency: in vitro adaptations of a continuum of in vivo states.BMC Dev Biol. 2017 Jun 13;17(1):7. doi: 10.1186/s12861-017-0150-4. BMC Dev Biol. 2017. PMID: 28610558 Free PMC article. Review.
-
Cited2, a transcriptional modulator protein, regulates metabolism in murine embryonic stem cells.J Biol Chem. 2014 Jan 3;289(1):251-63. doi: 10.1074/jbc.M113.497594. Epub 2013 Nov 21. J Biol Chem. 2014. PMID: 24265312 Free PMC article.
References
-
- Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., Vallier L. (2007) Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 - PubMed
-
- Evans M. J., Kaufman M. H. (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

