LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis
- PMID: 22822152
- PMCID: PMC3432903
- DOI: 10.1126/science.1220281
LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis
Abstract
Defective catabolite export from lysosomes results in lysosomal storage diseases in humans. Mutations in the cystine transporter gene CTNS cause cystinosis, but other lysosomal amino acid transporters are poorly characterized at the molecular level. Here, we identified the Caenorhabditis elegans lysosomal lysine/arginine transporter LAAT-1. Loss of laat-1 caused accumulation of lysine and arginine in enlarged, degradation-defective lysosomes. In mutants of ctns-1 (C. elegans homolog of CTNS), LAAT-1 was required to reduce lysosomal cystine levels and suppress lysosome enlargement by cysteamine, a drug that alleviates cystinosis by converting cystine to a lysine analog. LAAT-1 also maintained availability of cytosolic lysine/arginine during embryogenesis. Thus, LAAT-1 is the lysosomal lysine/arginine transporter, which suggests a molecular explanation for how cysteamine alleviates a lysosomal storage disease.
Figures

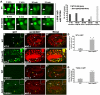
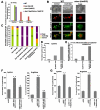
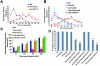
Similar articles
-
Heptahelical protein PQLC2 is a lysosomal cationic amino acid exporter underlying the action of cysteamine in cystinosis therapy.Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):E3434-43. doi: 10.1073/pnas.1211198109. Epub 2012 Nov 20. Proc Natl Acad Sci U S A. 2012. PMID: 23169667 Free PMC article.
-
Arginine-selective modulation of the lysosomal transporter PQLC2 through a gate-tuning mechanism.Proc Natl Acad Sci U S A. 2021 Aug 10;118(32):e2025315118. doi: 10.1073/pnas.2025315118. Proc Natl Acad Sci U S A. 2021. PMID: 34344826 Free PMC article.
-
Upregulation of the Rab27a-dependent trafficking and secretory mechanisms improves lysosomal transport, alleviates endoplasmic reticulum stress, and reduces lysosome overload in cystinosis.Mol Cell Biol. 2013 Aug;33(15):2950-62. doi: 10.1128/MCB.00417-13. Epub 2013 May 28. Mol Cell Biol. 2013. PMID: 23716592 Free PMC article.
-
Caenorhabditis elegans as a model for lysosomal storage disorders.Biochim Biophys Acta. 2008 Jul-Aug;1782(7-8):433-46. doi: 10.1016/j.bbadis.2008.04.003. Epub 2008 May 1. Biochim Biophys Acta. 2008. PMID: 18501720 Review.
-
Invertebrate models of lysosomal storage disease: what have we learned so far?Invert Neurosci. 2011 Dec;11(2):59-71. doi: 10.1007/s10158-011-0125-2. Epub 2011 Oct 25. Invert Neurosci. 2011. PMID: 22038288 Review.
Cited by
-
Functional Characterization of the Lysosomal Peptide/Histidine Transporter PHT1 (SLC15A4) by Solid Supported Membrane Electrophysiology (SSME).Biomolecules. 2024 Jun 28;14(7):771. doi: 10.3390/biom14070771. Biomolecules. 2024. PMID: 39062485 Free PMC article.
-
Role of transcription factors in apoptotic cells clearance.Front Cell Dev Biol. 2023 Jan 19;11:1110225. doi: 10.3389/fcell.2023.1110225. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36743409 Free PMC article. Review.
-
The lysosome as a cellular centre for signalling, metabolism and quality control.Nat Cell Biol. 2019 Feb;21(2):133-142. doi: 10.1038/s41556-018-0244-7. Epub 2019 Jan 2. Nat Cell Biol. 2019. PMID: 30602725 Review.
-
Lysosomal adaptation: how the lysosome responds to external cues.Cold Spring Harb Perspect Biol. 2014 May 5;6(6):a016907. doi: 10.1101/cshperspect.a016907. Cold Spring Harb Perspect Biol. 2014. PMID: 24799353 Free PMC article.
-
Spatiotemporal recruitment of the ubiquitin-specific protease USP8 directs endosome maturation.Elife. 2024 Nov 22;13:RP96353. doi: 10.7554/eLife.96353. Elife. 2024. PMID: 39576689 Free PMC article.
References
-
- Sagne C, Gasnier B. Molecular physiology and pathophysiology of lysosomal membrane transporters. J Inherit Metab Dis. 2008;31:258. - PubMed
-
- Gahl WA, Bashan N, Tietze F, Bernardini I, Schulman JD. Cystine transport is defective in isolated leukocyte lysosomes from patients with cystinosis. Science. 1982;217:1263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

