AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots
- PMID: 22807623
- PMCID: PMC3396566
- DOI: 10.4137/EBO.S9369
AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots
Abstract
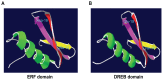
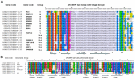
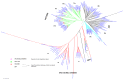
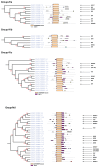
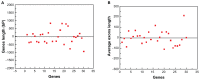
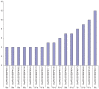
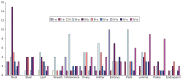
The transcription factor family intimately regulates gene expression in response to hormones, biotic and abiotic factors, symbiotic interactions, cell differentiation, and stress signalling pathways in plants. In this study, 170 AP2/ERF family genes are identified by phylogenetic analysis of the rice genome (Oryza sativa l. japonica) and they are divided into a total of 11 groups, including four major groups (AP2, ERF, DREB, and RAV), 10 subgroups, and two soloists. Gene structure analysis revealed that, at position-6, the amino acid threonine (Thr-6) is conserved in the double domain AP2 proteins compared to the amino acid arginine (Arg-6), which is preserved in the single domain of ERF proteins. In addition, the histidine (His) amino acid is found in both domains of the double domain AP2 protein, which is missing in single domain ERF proteins. Motif analysis indicates that most of the conserved motifs, apart from the AP2/ERF domain, are exclusively distributed among the specific clades in the phylogenetic tree and regulate plausible functions. Expression analysis reveals a widespread distribution of the rice AP2/ERF family genes within plant tissues. In the vegetative organs, the transcripts of these genes are found most abundant in the roots followed by the leaf and stem; whereas, in reproductive tissues, the gene expression of this family is observed high in the embryo and lemma. From chromosomal localization, it appears that repetition and tandem-duplication may contribute to the evolution of new genes in the rice genome. In this study, interspecies comparisons between rice and wheat reveal 34 rice loci and unveil the extent of collinearity between the two genomes. It was subsequently ascertained that chromosome-9 has more orthologous loci for CRT/DRE genes whereas chromosome-2 exhibits orthologs for ERF subfamily members. Maximum conserved synteny is found in chromosome-3 for AP2 double domain subfamily genes. Macrosynteny between rice and Arabidopsis, a distant, related genome, uncovered 11 homologs/orthologs loci in both genomes. The distribution of AP2/ERF family gene paralogs in Arabidopsis was most frequent in chromosome-1 followed by chromosome-5. In Arabidopsis, ERF subfamily gene orthologs are found on chromosome-1, chromosome-3, and chromosome-5, whereas DRE subfamily genes are found on chromosome-2 and chromosome-5. Orthologs for RAV and AP2 with double domains in Arabidopsis are located on chromosome-1 and chromosome-3, respectively. In conclusion, the data generated in this survey will be useful for conducting genomic research to determine the precise role of the AP2/ERF gene during stress responses with the ultimate goal of improving crops.
Keywords: AP2/ERF; Arabidopsis; CBF/DREB; RAV; phylogenetic analysis; rice; wheat.
Figures








Similar articles
-
Genome-wide identification, structural characterization and expression profiling of AP2/ERF gene family in bayberry (Myrica rubra).BMC Plant Biol. 2024 Nov 28;24(1):1139. doi: 10.1186/s12870-024-05847-2. BMC Plant Biol. 2024. PMID: 39604860 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
The effectiveness of school-based family asthma educational programs on the quality of life and number of asthma exacerbations of children aged five to 18 years diagnosed with asthma: a systematic review protocol.JBI Database System Rev Implement Rep. 2015 Oct;13(10):69-81. doi: 10.11124/jbisrir-2015-2335. JBI Database System Rev Implement Rep. 2015. PMID: 26571284
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
-
Comparative Transcriptome Analysis of High- and Low-Growth Genotypes of Eucalyptus urophylla in Response to Long-Term Nitrogen Deficiency.Genes (Basel). 2023 Dec 30;15(1):60. doi: 10.3390/genes15010060. Genes (Basel). 2023. PMID: 38254950 Free PMC article.
-
Genome-wide identification and expression analysis of the ERF transcription factor family in pineapple (Ananas comosus (L.) Merr.).PeerJ. 2020 Sep 22;8:e10014. doi: 10.7717/peerj.10014. eCollection 2020. PeerJ. 2020. PMID: 33024641 Free PMC article.
-
Genome-Wide Analysis of the AP2/ERF Transcription Factors Family and the Expression Patterns of DREB Genes in Moso Bamboo (Phyllostachys edulis).PLoS One. 2015 May 18;10(5):e0126657. doi: 10.1371/journal.pone.0126657. eCollection 2015. PLoS One. 2015. PMID: 25985202 Free PMC article.
-
Transcription Factor ERF194 Modulates the Stress-Related Physiology to Enhance Drought Tolerance of Poplar.Int J Mol Sci. 2023 Jan 2;24(1):788. doi: 10.3390/ijms24010788. Int J Mol Sci. 2023. PMID: 36614232 Free PMC article.
-
Wide-Range Portrayal of AP2/ERF Transcription Factor Family in Maize (Zea mays L.) Development and Stress Responses.Genes (Basel). 2023 Jan 11;14(1):194. doi: 10.3390/genes14010194. Genes (Basel). 2023. PMID: 36672935 Free PMC article.
References
-
- Abogadallah GM, Nada RM, Malinowski R, Quick P. Overexpression of HARDY, an AP2/ERF gene from Arabidopsis, improves drought and salt tolerance by reducing transpiration and sodium uptake in transgenic Trifolium alexandrinum L. Planta. 2011;233:1265–76. - PubMed
-
- Agarwal P, Agarwal PK, Joshi AJ, Sopory SK, Reddy MK. Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Molecular Biology Reports. 2010;37(2):1125–35. - PubMed
-
- Gao FC JM, Xiong AS, Peng RH, Liu JG, Cai B, Yao QH. Isolation and characterization of a novel AP2/EREBP-type transcription factor OsAP211 in Oryza sativa. Biologia Plantarum. 2009;53(4):643–9.
-
- Hu Y, Zhao L, Chong K, Wang T. Overexpression of OsERF1, a novel rice ERF gene, up-regulates ethylene-responsive genes expression besides affects growth and development in Arabidopsis. Journal of Plant Physiology. 2008;165(16):1717–25. - PubMed
-
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biological Chemistry. 1998;379:633–46. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

