Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis
- PMID: 22802657
- PMCID: PMC3412041
- DOI: 10.1073/pnas.1204915109
Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis
Abstract
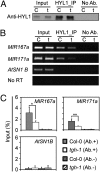
MicroRNAs (miRNAs) are regulators of gene expression in plants and animals. The biogenesis of miRNAs is precisely controlled to secure normal development of organisms. Here we report that TOUGH (TGH) is a component of the DCL1-HYL1-SERRATE complex that processes primary transcripts of miRNAs [i.e., primary miRNAs (pri-miRNAs)] into miRNAs in Arabidopsis. Lack of TGH impairs multiple DCL activities in vitro and reduces the accumulation of miRNAs and siRNAs in vivo. TGH is an RNA-binding protein, binds pri-miRNAs and precursor miRNAs in vivo, and contributes to pri-miRNA-HYL1 interaction. These results indicate that TGH might regulate abundance of miRNAs through promoting DCL1 cleavage efficiency and/or recruitment of pri-miRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
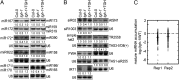
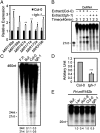
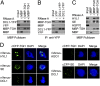
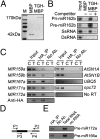
Similar articles
-
The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1.Proc Natl Acad Sci U S A. 2008 Jul 22;105(29):9970-5. doi: 10.1073/pnas.0803356105. Epub 2008 Jul 16. Proc Natl Acad Sci U S A. 2008. PMID: 18632569 Free PMC article.
-
Critical roles of RNA-binding proteins in miRNA biogenesis in Arabidopsis.RNA Biol. 2012 Dec;9(12):1424-8. doi: 10.4161/rna.22740. Epub 2012 Nov 7. RNA Biol. 2012. PMID: 23135480
-
Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA.Nucleic Acids Res. 2014 Oct 29;42(19):12224-36. doi: 10.1093/nar/gku907. Epub 2014 Oct 7. Nucleic Acids Res. 2014. PMID: 25294831 Free PMC article.
-
New insights into pri-miRNA processing and accumulation in plants.Wiley Interdiscip Rev RNA. 2015 Sep-Oct;6(5):533-45. doi: 10.1002/wrna.1292. Epub 2015 Jun 29. Wiley Interdiscip Rev RNA. 2015. PMID: 26119101 Review.
-
SERRATE: a key factor in coordinated RNA processing in plants.Trends Plant Sci. 2023 Jul;28(7):841-853. doi: 10.1016/j.tplants.2023.03.009. Epub 2023 Apr 4. Trends Plant Sci. 2023. PMID: 37019716 Review.
Cited by
-
Uridylation and the SKI complex orchestrate the Calvin cycle of photosynthesis through RNA surveillance of TKL1 in Arabidopsis.Proc Natl Acad Sci U S A. 2022 Sep 20;119(38):e2205842119. doi: 10.1073/pnas.2205842119. Epub 2022 Sep 12. Proc Natl Acad Sci U S A. 2022. PMID: 36095196 Free PMC article.
-
Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3' to 5' exoribonuclease Atrimmer 2 in Arabidopsis.Proc Natl Acad Sci U S A. 2018 Jul 10;115(28):E6659-E6667. doi: 10.1073/pnas.1721917115. Epub 2018 Jun 25. Proc Natl Acad Sci U S A. 2018. PMID: 29941559 Free PMC article.
-
The spliceosome-associated protein CWC15 promotes miRNA biogenesis in Arabidopsis.Nat Commun. 2024 Mar 16;15(1):2399. doi: 10.1038/s41467-024-46676-z. Nat Commun. 2024. PMID: 38493158 Free PMC article.
-
Sugar Signaling and Post-transcriptional Regulation in Plants: An Overlooked or an Emerging Topic?Front Plant Sci. 2020 Nov 5;11:578096. doi: 10.3389/fpls.2020.578096. eCollection 2020. Front Plant Sci. 2020. PMID: 33224165 Free PMC article. Review.
-
Identification of RNA-based cell-type markers for stem-cell manufacturing systems with a statistical scoring function.Gene Rep. 2024 Mar;34:101869. doi: 10.1016/j.genrep.2023.101869. Epub 2023 Dec 12. Gene Rep. 2024. PMID: 38351912 Free PMC article.
References
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. - PubMed
-
- Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48:1243–1253. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

