The faah gene is the first direct target of estrogen in the testis: role of histone demethylase LSD1
- PMID: 22802127
- PMCID: PMC11114663
- DOI: 10.1007/s00018-012-1074-6
The faah gene is the first direct target of estrogen in the testis: role of histone demethylase LSD1
Abstract
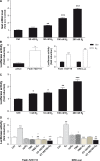
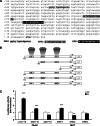
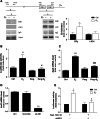
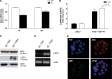
Estrogen (E(2)) regulates spermatogenesis, yet its direct target genes have not been identified in the testis. Here, we cloned the proximal 5' flanking region of the mouse fatty acid amide hydrolase (faah) gene upstream of the luciferase reporter gene, and demonstrated its promoter activity and E(2) inducibility in primary mouse Sertoli cells. Specific mutations in the E(2) response elements (ERE) of the faah gene showed that two proximal ERE sequences (ERE2/3) are essential for E(2)-induced transcription, and chromatin immunoprecipitation experiments showed that E(2) induced estrogen receptor β binding at ERE2/3 sites in the faah promoter in vivo. Moreover, the histone demethylase LSD1 was found to be associated with ERE2/3 sites and to play a role in mediating E(2) induction of FAAH expression. E(2) induced epigenetic modifications at the faah proximal promoter compatible with transcriptional activation by remarkably decreasing methylation of both DNA at CpG site and histone H3 at lysine 9. Finally, FAAH silencing abolished E(2) protection against apoptosis induced by the FAAH substrate anandamide. Taken together, our results identify FAAH as the first direct target of E(2).
Figures

Similar articles
-
Histone lysine-specific demethylase 1 (LSD1) protein is involved in Sal-like protein 4 (SALL4)-mediated transcriptional repression in hematopoietic stem cells.J Biol Chem. 2013 Nov 29;288(48):34719-28. doi: 10.1074/jbc.M113.506568. Epub 2013 Oct 25. J Biol Chem. 2013. PMID: 24163373 Free PMC article.
-
Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression.Nature. 2007 Aug 9;448(7154):718-22. doi: 10.1038/nature06034. Nature. 2007. PMID: 17687328 Free PMC article.
-
LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription.Nature. 2005 Sep 15;437(7057):436-9. doi: 10.1038/nature04020. Epub 2005 Aug 3. Nature. 2005. PMID: 16079795
-
New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin.FEBS J. 2009 Aug;276(16):4304-12. doi: 10.1111/j.1742-4658.2009.07142.x. Epub 2009 Jul 14. FEBS J. 2009. PMID: 19624733 Review.
-
Mechanisms involved in the regulation of histone lysine demethylases.Curr Opin Cell Biol. 2008 Jun;20(3):316-25. doi: 10.1016/j.ceb.2008.03.004. Epub 2008 Apr 25. Curr Opin Cell Biol. 2008. PMID: 18440794 Free PMC article. Review.
Cited by
-
Environmental epigenetics and effects on male fertility.Adv Exp Med Biol. 2014;791:67-81. doi: 10.1007/978-1-4614-7783-9_5. Adv Exp Med Biol. 2014. PMID: 23955673 Free PMC article. Review.
-
Behavioral effects and mechanisms of migraine pathogenesis following estradiol exposure in a multibehavioral model of migraine in rat.Exp Neurol. 2015 Jan;263:8-16. doi: 10.1016/j.expneurol.2014.09.011. Epub 2014 Sep 28. Exp Neurol. 2015. PMID: 25263582 Free PMC article.
-
Fatty acid amide hydrolase drives adult mammary gland development by promoting luminal cell differentiation.Cell Death Discov. 2024 Jan 6;10(1):12. doi: 10.1038/s41420-023-01788-1. Cell Death Discov. 2024. PMID: 38184644 Free PMC article.
-
Updates in reproduction coming from the endocannabinoid system.Int J Endocrinol. 2014;2014:412354. doi: 10.1155/2014/412354. Epub 2014 Jan 16. Int J Endocrinol. 2014. PMID: 24550985 Free PMC article. Review.
-
Quality of Life and a Surveillant Endocannabinoid System.Front Neurosci. 2021 Oct 28;15:747229. doi: 10.3389/fnins.2021.747229. eCollection 2021. Front Neurosci. 2021. PMID: 34776851 Free PMC article. Review.
References
-
- Zhou Q, Nie R, Prins GS, Saunders PT, Katzenellenbogen BS, Hess RA. Localization of androgen and estrogen receptors in adult male mouse reproductive tract. J Androl. 2002;23:870–881. - PubMed
-
- Lucas TF, Siu ER, Esteves CA, Monteiro HP, Oliveira CA, Porto CS, Lazari MF. 17beta-estradiol induces the translocation of the estrogen receptors ESR1 and ESR2 to the cell membrane, MAPK3/1 phosphorylation and proliferation of cultured immature rat Sertoli cells. Biol Reprod. 2008;78:101–114. doi: 10.1095/biolreprod.107.063909. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

