Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant
- PMID: 22798321
- PMCID: PMC3480531
- DOI: 10.1164/rccm.201204-0693OC
Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant
Abstract
Rationale: Long-term survival after lung transplantation is limited by infectious complications and by bronchiolitis obliterans syndrome (BOS), a form of chronic rejection linked in part to microbial triggers.
Objectives: To define microbial populations in the respiratory tract of transplant patients comprehensively using unbiased high-density sequencing.
Methods: Lung was sampled by bronchoalveolar lavage (BAL) and upper respiratory tract by oropharyngeal wash (OW). Bacterial 16S rDNA and fungal internal transcribed spacer sequencing was used to profile organisms present. Outlier analysis plots defining taxa enriched in lung relative to OW were used to identify bacteria enriched in lung against a background of oropharyngeal carryover.
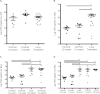
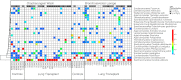
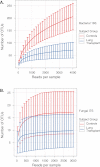
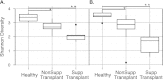
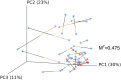
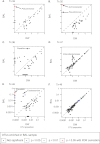
Measurements and main results: Lung transplant recipients had higher bacterial burden in BAL than control subjects, frequent appearance of dominant organisms, greater distance between communities in BAL and OW indicating more distinct populations, and decreased respiratory tract microbial richness and diversity. Fungal populations were typically dominated by Candida in both sites or by Aspergillus in BAL but not OW. 16S outlier analysis identified lung-enriched taxa indicating bacteria replicating in the lower respiratory tract. In some cases this confirmed respiratory cultures but in others revealed enrichment by anaerobic organisms or mixed outgrowth of upper respiratory flora and provided quantitative data on relative abundances of bacteria found by culture.
Conclusions: Respiratory tract microbial communities in lung transplant recipients differ in structure and composition from healthy subjects. Outlier analysis can identify specific bacteria replicating in lung. These findings provide novel approaches to address the relationship between microbial communities and transplant outcome and aid in assessing lung infections.
Figures

Similar articles
-
Upper Respiratory Dysbiosis with a Facultative-dominated Ecotype in Advanced Lung Disease and Dynamic Change after Lung Transplant.Ann Am Thorac Soc. 2019 Nov;16(11):1383-1391. doi: 10.1513/AnnalsATS.201904-299OC. Ann Am Thorac Soc. 2019. PMID: 31415219 Free PMC article.
-
Reestablishment of recipient-associated microbiota in the lung allograft is linked to reduced risk of bronchiolitis obliterans syndrome.Am J Respir Crit Care Med. 2013 Mar 15;187(6):640-7. doi: 10.1164/rccm.201209-1680OC. Epub 2013 Jan 17. Am J Respir Crit Care Med. 2013. PMID: 23328523
-
Effect of etiology and timing of respiratory tract infections on development of bronchiolitis obliterans syndrome.J Heart Lung Transplant. 2009 Feb;28(2):163-9. doi: 10.1016/j.healun.2008.11.907. J Heart Lung Transplant. 2009. PMID: 19201342
-
[Infection in lung transplantation].Enferm Infecc Microbiol Clin. 2007 Dec;25(10):639-49; quiz 650. doi: 10.1157/13112940. Enferm Infecc Microbiol Clin. 2007. PMID: 18053475 Free PMC article. Review. Spanish.
-
The lung microbiome after lung transplantation.Expert Rev Respir Med. 2014 Apr;8(2):221-31. doi: 10.1586/17476348.2014.890518. Expert Rev Respir Med. 2014. PMID: 24601662 Free PMC article. Review.
Cited by
-
Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota.Microbiome. 2016 Jun 23;4(1):29. doi: 10.1186/s40168-016-0172-3. Microbiome. 2016. PMID: 27338728 Free PMC article.
-
Unraveling the gut-Lung axis: Exploring complex mechanisms in disease interplay.Heliyon. 2024 Jan 3;10(1):e24032. doi: 10.1016/j.heliyon.2024.e24032. eCollection 2024 Jan 15. Heliyon. 2024. PMID: 38268584 Free PMC article. Review.
-
An Update in Antimicrobial Therapies and Infection Prevention in Pediatric Lung Transplant Recipients.Paediatr Drugs. 2018 Dec;20(6):539-553. doi: 10.1007/s40272-018-0313-1. Paediatr Drugs. 2018. PMID: 30187362 Review.
-
Lack of detection of a human placenta microbiome in samples from preterm and term deliveries.Microbiome. 2018 Oct 30;6(1):196. doi: 10.1186/s40168-018-0575-4. Microbiome. 2018. PMID: 30376898 Free PMC article.
-
Recent Advances in Fungal Infections: From Lung Ecology to Therapeutic Strategies With a Focus on Aspergillus spp.Front Med (Lausanne). 2022 Mar 21;9:832510. doi: 10.3389/fmed.2022.832510. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35386908 Free PMC article. Review.
References
-
- Christie JD, Edwards LB, Kucheryavaya AY, Benden C, Dobbels F, Kirk R, Rahmel AO, Stehlik J, Hertz MI. The registry of the international society for heart and lung transplantation: twenty-eighth adult lung and heart-lung transplant report–2011. J Heart Lung Transplant 2011;30:1104–1122 - PubMed
-
- Robertson AG, Griffin SM, Murphy DM, Pearson JP, Forrest IA, Dark JH, Corris PA, Ward C. Targeting allograft injury and inflammation in the management of post-lung transplant bronchiolitis obliterans syndrome. Am J Transplant 2009;9:1272–1278 - PubMed
-
- Atkins BZ, Trachtenberg MS, Prince-Petersen R, Vess G, Bush EL, Balsara KR, Lin SS, Davis RD., Jr Assessing oropharyngeal dysphagia after lung transplantation: altered swallowing mechanisms and increased morbidity. J Heart Lung Transplant 2007;26:1144–1148 - PubMed
-
- Botha P, Archer L, Anderson RL, Lordan J, Dark JH, Corris PA, Gould K, Fisher AJ. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation 2008;85:771–774 - PubMed
-
- Khalifah AP, Hachem RR, Chakinala MM, Schechtman KB, Patterson GA, Schuster DP, Mohanakumar T, Trulock EP, Walter MJ. Respiratory viral infections are a distinct risk for bronchiolitis obliterans syndrome and death. Am J Respir Crit Care Med 2004;170:181–187 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

