Role of architecture in the function and specificity of two Notch-regulated transcriptional enhancer modules
- PMID: 22792075
- PMCID: PMC3390367
- DOI: 10.1371/journal.pgen.1002796
Role of architecture in the function and specificity of two Notch-regulated transcriptional enhancer modules
Abstract
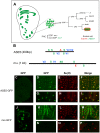
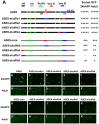
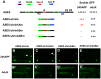
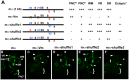
In Drosophila melanogaster, cis-regulatory modules that are activated by the Notch cell-cell signaling pathway all contain two types of transcription factor binding sites: those for the pathway's transducing factor Suppressor of Hairless [Su(H)] and those for one or more tissue- or cell type-specific factors called "local activators." The use of different "Su(H) plus local activator" motif combinations, or codes, is critical to ensure that only the correct subset of the broadly utilized Notch pathway's target genes are activated in each developmental context. However, much less is known about the role of enhancer "architecture"--the number, order, spacing, and orientation of its component transcription factor binding motifs--in determining the module's specificity. Here we investigate the relationship between architecture and function for two Notch-regulated enhancers with spatially distinct activities, each of which includes five high-affinity Su(H) sites. We find that the first, which is active specifically in the socket cells of external sensory organs, is largely resistant to perturbations of its architecture. By contrast, the second enhancer, active in the "non-SOP" cells of the proneural clusters from which neural precursors arise, is sensitive to even simple rearrangements of its transcription factor binding sites, responding with both loss of normal specificity and striking ectopic activity. Thus, diverse cryptic specificities can be inherent in an enhancer's particular combination of transcription factor binding motifs. We propose that for certain types of enhancer, architecture plays an essential role in determining specificity, not only by permitting factor-factor synergies necessary to generate the desired activity, but also by preventing other activator synergies that would otherwise lead to unwanted specificities.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Three distinct mechanisms, Notch instructive, permissive, and independent, regulate the expression of two different pericardial genes to specify cardiac cell subtypes.PLoS One. 2020 Oct 27;15(10):e0241191. doi: 10.1371/journal.pone.0241191. eCollection 2020. PLoS One. 2020. PMID: 33108408 Free PMC article.
-
Molecular analysis of the notch repressor-complex in Drosophila: characterization of potential hairless binding sites on suppressor of hairless.PLoS One. 2011;6(11):e27986. doi: 10.1371/journal.pone.0027986. Epub 2011 Nov 18. PLoS One. 2011. PMID: 22125648 Free PMC article.
-
Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless.Development. 2005 Aug;132(15):3333-44. doi: 10.1242/dev.01920. Epub 2005 Jun 23. Development. 2005. PMID: 15975935
-
Notch pathway: making sense of suppressor of hairless.Curr Biol. 2001 Mar 20;11(6):R217-21. doi: 10.1016/s0960-9822(01)00109-9. Curr Biol. 2001. PMID: 11301266 Review.
-
Sparkling insights into enhancer structure, function, and evolution.Curr Top Dev Biol. 2012;98:97-120. doi: 10.1016/B978-0-12-386499-4.00004-5. Curr Top Dev Biol. 2012. PMID: 22305160 Review.
Cited by
-
Enhancer evolution and the origins of morphological novelty.Curr Opin Genet Dev. 2017 Aug;45:115-123. doi: 10.1016/j.gde.2017.04.006. Epub 2017 May 18. Curr Opin Genet Dev. 2017. PMID: 28527813 Free PMC article. Review.
-
Cis-regulatory complexity within a large non-coding region in the Drosophila genome.PLoS One. 2013 Apr 22;8(4):e60137. doi: 10.1371/journal.pone.0060137. Print 2013. PLoS One. 2013. PMID: 23613719 Free PMC article.
-
Assessing constraints on the path of regulatory sequence evolution.Philos Trans R Soc Lond B Biol Sci. 2013 Nov 11;368(1632):20130026. doi: 10.1098/rstb.2013.0026. Print 2013 Dec 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24218638 Free PMC article.
-
The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force.Dev Cell. 2017 May 8;41(3):228-241. doi: 10.1016/j.devcel.2017.04.001. Dev Cell. 2017. PMID: 28486129 Free PMC article. Review.
-
Enhancer grammar in development, evolution, and disease: dependencies and interplay.Dev Cell. 2021 Mar 8;56(5):575-587. doi: 10.1016/j.devcel.2021.02.016. Dev Cell. 2021. PMID: 33689769 Free PMC article. Review.
References
-
- Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. - PubMed
-
- Davidson EH. The regulatory genome. Gene regulatory networks in development and evolution. San Diego: Academic Press; 2006.
-
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. - PubMed
-
- Crocker J, Erives A. A closer look at the eve stripe 2 enhancers of Drosophila and Themira. PLoS Genet. 2008;4:e1000276. doi: 10.1371/journal.pgen.1000276. - DOI - PMC - PubMed
-
- Hare EE, Peterson BK, Eisen MB. A careful look at binding site reorganization in the even-skipped enhancers of Drosophila and sepsids. PLoS Genet. 2008;4:e1000268. doi: 10.1371/journal.pgen.1000268. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

