miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis
- PMID: 22772450
- PMCID: PMC3411895
- DOI: 10.4049/jimmunol.1103171
miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis
Abstract
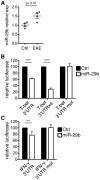
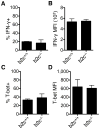
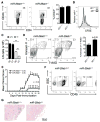
Th cell programming and function is tightly regulated by complex biological networks to prevent excessive inflammatory responses and autoimmune disease. The importance of microRNAs (miRNAs) in this process is highlighted by the preferential Th1 polarization of Dicer-deficient T cells that lack miRNAs. Using genetic knockouts, we demonstrate that loss of endogenous miR-29, derived from the miR-29ab1 genomic cluster, results in unrestrained T-bet expression and IFN-γ production. miR-29b regulates T-bet and IFN-γ via a direct interaction with the 3' untranslated regions, and IFN-γ itself enhances miR-29b expression, establishing a novel regulatory feedback loop. miR-29b is increased in memory CD4(+) T cells from multiple sclerosis (MS) patients, which may reflect chronic Th1 inflammation. However, miR-29b levels decrease significantly upon T cell activation in MS patients, suggesting that this feedback loop is dysregulated in MS patients and may contribute to chronic inflammation. miR-29 thus serves as a novel regulator of Th1 differentiation, adding to the understanding of T cell-intrinsic regulatory mechanisms that maintain a balance between protective immunity and autoimmunity.
Figures

Similar articles
-
miR-17-92 cluster targets phosphatase and tensin homology and Ikaros Family Zinc Finger 4 to promote TH17-mediated inflammation.J Biol Chem. 2014 May 2;289(18):12446-56. doi: 10.1074/jbc.M114.550723. Epub 2014 Mar 18. J Biol Chem. 2014. PMID: 24644282 Free PMC article.
-
MicroRNA223 promotes pathogenic T-cell development and autoimmune inflammation in central nervous system in mice.Immunology. 2016 Aug;148(4):326-38. doi: 10.1111/imm.12611. Epub 2016 Jun 29. Immunology. 2016. PMID: 27083389 Free PMC article.
-
miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORγt and STAT3.J Immunol. 2014 Jun 15;192(12):5599-609. doi: 10.4049/jimmunol.1303488. Epub 2014 May 19. J Immunol. 2014. PMID: 24842756
-
MicroRNAs and their Implications in CD4+ T-cells, Oligodendrocytes and Dendritic Cells in Multiple Sclerosis Pathogenesis.Curr Mol Med. 2023 May 30;23(7):630-647. doi: 10.2174/1566524022666220525150259. Curr Mol Med. 2023. PMID: 35619281 Review.
-
Dysregulated Network of miRNAs Involved in the Pathogenesis of Multiple Sclerosis.Biomed Pharmacother. 2018 Aug;104:280-290. doi: 10.1016/j.biopha.2018.05.050. Epub 2018 May 25. Biomed Pharmacother. 2018. PMID: 29775896 Review.
Cited by
-
Control of the Inflammatory Macrophage Transcriptional Signature by miR-155.PLoS One. 2016 Jul 22;11(7):e0159724. doi: 10.1371/journal.pone.0159724. eCollection 2016. PLoS One. 2016. PMID: 27447824 Free PMC article.
-
Role of miRNAs in CD4 T cell plasticity during inflammation and tolerance.Front Genet. 2013 Jan 31;4:8. doi: 10.3389/fgene.2013.00008. eCollection 2013. Front Genet. 2013. PMID: 23386861 Free PMC article.
-
The inflammatory effect of epigenetic factors and modifications in type 2 diabetes.Inflammopharmacology. 2020 Apr;28(2):345-362. doi: 10.1007/s10787-019-00663-9. Epub 2019 Nov 9. Inflammopharmacology. 2020. PMID: 31707555 Review.
-
Investigating the Role of MicroRNA and Transcription Factor Co-regulatory Networks in Multiple Sclerosis Pathogenesis.Int J Mol Sci. 2018 Nov 20;19(11):3652. doi: 10.3390/ijms19113652. Int J Mol Sci. 2018. PMID: 30463275 Free PMC article.
-
Posttranscriptional regulation of T helper cell fate decisions.J Cell Biol. 2018 Aug 6;217(8):2615-2631. doi: 10.1083/jcb.201708075. Epub 2018 Apr 23. J Cell Biol. 2018. PMID: 29685903 Free PMC article. Review.
References
-
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. - PubMed
-
- Voskuhl RR, Martin R, Bergman C, Dalal M, Ruddle NH, McFarland HF. T helper 1 (Th1) functional phenotype of human myelin basic protein-specific T lymphocytes. Autoimmunity. 1993;15:137–143. - PubMed
-
- Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, Langer-Gould A, Strober S, Cannella B, Allard J, Klonowski P, Austin A, Lad N, Kaminski N, Galli SJ, Oksenberg JR, Raine CS, Heller R, Steinman L. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med. 2002;8:500–508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000090/TR/NCATS NIH HHS/United States
- R21 NS067383/NS/NINDS NIH HHS/United States
- R01 NS067441/NS/NINDS NIH HHS/United States
- TL1RR025753/RR/NCRR NIH HHS/United States
- K24 NS044250/NS/NINDS NIH HHS/United States
- K24 NS44250/NS/NINDS NIH HHS/United States
- R21 AI092417/AI/NIAID NIH HHS/United States
- R01 AI43376/AI/NIAID NIH HHS/United States
- T32 GM068412/GM/NIGMS NIH HHS/United States
- R01 AI064320/AI/NIAID NIH HHS/United States
- R01 NS037513/NS/NINDS NIH HHS/United States
- TL1 RR025753/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

