Histone H3 lysine 4 methylation marks postreplicative human cytomegalovirus chromatin
- PMID: 22761369
- PMCID: PMC3446588
- DOI: 10.1128/JVI.00581-12
Histone H3 lysine 4 methylation marks postreplicative human cytomegalovirus chromatin
Abstract
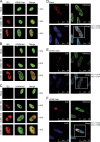
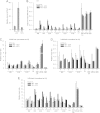
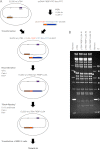
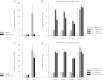
In the nuclei of permissive cells, human cytomegalovirus genomes form nucleosomal structures initially resembling heterochromatin but gradually switching to a euchromatin-like state. This switch is characterized by a decrease in histone H3 K9 methylation and a marked increase in H3 tail acetylation and H3 K4 methylation across the viral genome. We used ganciclovir and a mutant virus encoding a reversibly destabilized DNA polymerase to examine the impact of DNA replication on histone modification dynamics at the viral chromatin. The changes in H3 tail acetylation and H3 K9 methylation proceeded in a DNA replication-independent fashion. In contrast, the increase in H3 K4 methylation proved to depend widely on viral DNA synthesis. Consistently, labeling of nascent DNA using "click chemistry" revealed preferential incorporation of methylated H3 K4 into viral (but not cellular) chromatin during or following DNA replication. This study demonstrates largely selective epigenetic tagging of postreplicative human cytomegalovirus chromatin.
Figures

Similar articles
-
Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression.Mol Cell Biol. 2006 Sep;26(17):6357-71. doi: 10.1128/MCB.00311-06. Mol Cell Biol. 2006. PMID: 16914722 Free PMC article.
-
Histone modifications in Arabidopsis- high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin.Plant J. 2003 Feb;33(3):471-80. doi: 10.1046/j.1365-313x.2003.01638.x. Plant J. 2003. PMID: 12581305
-
Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content.Plant J. 2003 Mar;33(6):967-73. doi: 10.1046/j.1365-313x.2003.01681.x. Plant J. 2003. PMID: 12631322
-
Acetylation of yeast histone H4 lysine 16: a switch for protein interactions in heterochromatin and euchromatin.Cold Spring Harb Symp Quant Biol. 2004;69:193-200. doi: 10.1101/sqb.2004.69.193. Cold Spring Harb Symp Quant Biol. 2004. PMID: 16117649 Review. No abstract available.
-
[On the research of histone methylation].Yi Chuan. 2004 Mar;26(2):244-8. Yi Chuan. 2004. PMID: 15639996 Review. Chinese.
Cited by
-
Myeloblastic cell lines mimic some but not all aspects of human cytomegalovirus experimental latency defined in primary CD34+ cell populations.J Virol. 2013 Sep;87(17):9802-12. doi: 10.1128/JVI.01436-13. Epub 2013 Jul 3. J Virol. 2013. PMID: 23824798 Free PMC article.
-
Role of Epitranscriptomic and Epigenetic Modifications during the Lytic and Latent Phases of Herpesvirus Infections.Microorganisms. 2022 Aug 30;10(9):1754. doi: 10.3390/microorganisms10091754. Microorganisms. 2022. PMID: 36144356 Free PMC article. Review.
-
An Essential Viral Transcription Activator Modulates Chromatin Dynamics.PLoS Pathog. 2016 Aug 30;12(8):e1005842. doi: 10.1371/journal.ppat.1005842. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27575707 Free PMC article.
-
Regulation of the MIE Locus During HCMV Latency and Reactivation.Pathogens. 2020 Oct 23;9(11):869. doi: 10.3390/pathogens9110869. Pathogens. 2020. PMID: 33113934 Free PMC article. Review.
-
Human cytomegalovirus major immediate early 1 protein targets host chromosomes by docking to the acidic pocket on the nucleosome surface.J Virol. 2014 Jan;88(2):1228-48. doi: 10.1128/JVI.02606-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227840 Free PMC article.
References
-
- Barth TK, Imhof A. 2010. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 35:618–626 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

