Cyclophilins facilitate dissociation of the human papillomavirus type 16 capsid protein L1 from the L2/DNA complex following virus entry
- PMID: 22761365
- PMCID: PMC3446629
- DOI: 10.1128/JVI.00980-12
Cyclophilins facilitate dissociation of the human papillomavirus type 16 capsid protein L1 from the L2/DNA complex following virus entry
Abstract
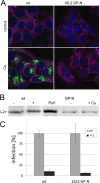
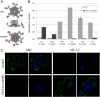
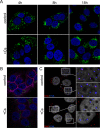
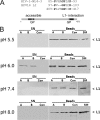
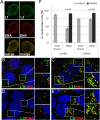
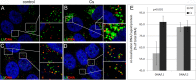
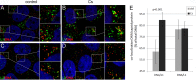
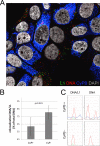
Human papillomaviruses (HPV) are composed of the major and minor capsid proteins, L1 and L2, that encapsidate a chromatinized, circular double-stranded DNA genome. At the outset of infection, the interaction of HPV type 16 (HPV16) (pseudo)virions with heparan sulfate proteoglycans triggers a conformational change in L2 that is facilitated by the host cell chaperone cyclophilin B (CyPB). This conformational change results in exposure of the L2 N terminus, which is required for infectious internalization. Following internalization, L2 facilitates egress of the viral genome from acidified endosomes, and the L2/DNA complex accumulates at PML nuclear bodies. We recently described a mutant virus that bypasses the requirement for cell surface CyPB but remains sensitive to cyclosporine for infection, indicating an additional role for CyP following endocytic uptake of virions. We now report that the L1 protein dissociates from the L2/DNA complex following infectious internalization. Inhibition and small interfering RNA (siRNA)-mediated knockdown of CyPs blocked dissociation of L1 from the L2/DNA complex. In vitro, purified CyPs facilitated the dissociation of L1 pentamers from recombinant HPV11 L1/L2 complexes in a pH-dependent manner. Furthermore, CyPs released L1 capsomeres from partially disassembled HPV16 pseudovirions at slightly acidic pH. Taken together, these data suggest that CyPs mediate the dissociation of HPV L1 and L2 capsid proteins following acidification of endocytic vesicles.
Figures
Similar articles
-
Target cell cyclophilins facilitate human papillomavirus type 16 infection.PLoS Pathog. 2009 Jul;5(7):e1000524. doi: 10.1371/journal.ppat.1000524. Epub 2009 Jul 24. PLoS Pathog. 2009. PMID: 19629175 Free PMC article.
-
Phosphorylation of Human Papillomavirus Type 16 L2 Contributes to Efficient Virus Infectious Entry.J Virol. 2019 Jun 14;93(13):e00128-19. doi: 10.1128/JVI.00128-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996086 Free PMC article.
-
The Cytoskeletal Adaptor Obscurin-Like 1 Interacts with the Human Papillomavirus 16 (HPV16) Capsid Protein L2 and Is Required for HPV16 Endocytosis.J Virol. 2016 Nov 14;90(23):10629-10641. doi: 10.1128/JVI.01222-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27654294 Free PMC article.
-
L2, the minor capsid protein of papillomavirus.Virology. 2013 Oct;445(1-2):175-86. doi: 10.1016/j.virol.2013.04.017. Epub 2013 May 17. Virology. 2013. PMID: 23689062 Free PMC article. Review.
-
The papillomavirus major capsid protein L1.Virology. 2013 Oct;445(1-2):169-74. doi: 10.1016/j.virol.2013.05.038. Epub 2013 Jun 22. Virology. 2013. PMID: 23800545 Free PMC article. Review.
Cited by
-
Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection.J Virol. 2013 Apr;87(7):3862-70. doi: 10.1128/JVI.03222-12. Epub 2013 Jan 23. J Virol. 2013. PMID: 23345514 Free PMC article.
-
The Interplay between Antiviral Signalling and Carcinogenesis in Human Papillomavirus Infections.Cancers (Basel). 2020 Mar 10;12(3):646. doi: 10.3390/cancers12030646. Cancers (Basel). 2020. PMID: 32164347 Free PMC article. Review.
-
Subcellular Trafficking of the Papillomavirus Genome during Initial Infection: The Remarkable Abilities of Minor Capsid Protein L2.Viruses. 2017 Dec 3;9(12):370. doi: 10.3390/v9120370. Viruses. 2017. PMID: 29207511 Free PMC article. Review.
-
A Dual Role for the Nonreceptor Tyrosine Kinase Pyk2 during the Intracellular Trafficking of Human Papillomavirus 16.J Virol. 2015 Sep;89(17):9103-14. doi: 10.1128/JVI.01183-15. Epub 2015 Jun 24. J Virol. 2015. PMID: 26109718 Free PMC article.
-
The evolving field of human papillomavirus receptor research: a review of binding and entry.J Virol. 2013 Jun;87(11):6062-72. doi: 10.1128/JVI.00330-13. Epub 2013 Mar 27. J Virol. 2013. PMID: 23536685 Free PMC article. Review.
References
-
- Bienkowska-Haba M, Patel HD, Sapp M. 2009. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 5:e1000524 doi:10.1371/journal.ppat.1000524 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

