Energy landscape and multiroute folding of topologically complex proteins adenylate kinase and 2ouf-knot
- PMID: 22753508
- PMCID: PMC3497823
- DOI: 10.1073/pnas.1201807109
Energy landscape and multiroute folding of topologically complex proteins adenylate kinase and 2ouf-knot
Erratum in
- Proc Natl Acad Sci U S A. 2012 Nov 6;109(45):18625
Abstract
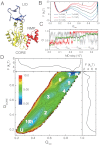
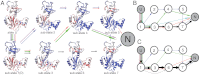
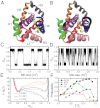
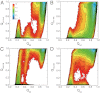
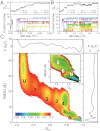
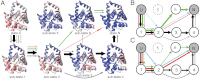
While fast folding of small proteins has been relatively well characterized by experiments and theories, much less is known for slow folding of larger proteins, for which recent experiments suggested quite complex and rich folding behaviors. Here, we address how the energy landscape theory can be applied to these slow folding reactions. Combining the perfect-funnel approximation with a multiscale method, we first extended our previous atomic-interaction based coarse grained (AICG) model to take into account local flexibility of protein molecules. Using this model, we then investigated the energy landscapes and folding routes of two proteins with complex topologies: a multidomain protein adenylate kinase (AKE) and a knotted protein 2ouf-knot. In the AKE folding, consistent with experimental results, the kinetic free energy surface showed several substates between the fully unfolded and native states. We characterized the structural features of these substates and transitions among them, finding temperature-dependent multiroute folding. For protein 2ouf-knot, we found that the improved atomic-interaction based coarse-grained model can spontaneously tie a knot and fold the protein with a probability up to 96%. The computed folding rate of the knotted protein was much slower than that of its unknotted counterpart, in agreement with experimental findings. Similar to the AKE case, the 2ouf-knot folding exhibited several substates and transitions among them. Interestingly, we found a dead-end substate that lacks the knot, thus suggesting backtracking mechanisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Energy landscape of knotted protein folding.Proc Natl Acad Sci U S A. 2012 Oct 30;109(44):17783-8. doi: 10.1073/pnas.1201804109. Epub 2012 Aug 13. Proc Natl Acad Sci U S A. 2012. PMID: 22891304 Free PMC article.
-
In the multi-domain protein adenylate kinase, domain insertion facilitates cooperative folding while accommodating function at domain interfaces.PLoS Comput Biol. 2014 Nov 13;10(11):e1003938. doi: 10.1371/journal.pcbi.1003938. eCollection 2014 Nov. PLoS Comput Biol. 2014. PMID: 25393408 Free PMC article.
-
Protein folding pathways of adenylate kinase from E. coli: hydrostatic pressure and stopped-flow studies.Biochemistry. 2001 Dec 4;40(48):14706-14. doi: 10.1021/bi010308i. Biochemistry. 2001. PMID: 11724585
-
Elucidation of folding pathways of knotted proteins.Methods Enzymol. 2022;675:275-297. doi: 10.1016/bs.mie.2022.07.012. Epub 2022 Aug 19. Methods Enzymol. 2022. PMID: 36220273 Review.
-
Insights from coarse-grained Gō models for protein folding and dynamics.Int J Mol Sci. 2009 Mar;10(3):889-905. doi: 10.3390/ijms10030889. Epub 2009 Mar 2. Int J Mol Sci. 2009. PMID: 19399227 Free PMC article. Review.
Cited by
-
Investigating the trade-off between folding and function in a multidomain Y-family DNA polymerase.Elife. 2020 Oct 20;9:e60434. doi: 10.7554/eLife.60434. Elife. 2020. PMID: 33079059 Free PMC article.
-
How Co-translational Folding of Multi-domain Protein Is Affected by Elongation Schedule: Molecular Simulations.PLoS Comput Biol. 2015 Jul 9;11(7):e1004356. doi: 10.1371/journal.pcbi.1004356. eCollection 2015 Jul. PLoS Comput Biol. 2015. PMID: 26158498 Free PMC article.
-
Cooperativity and Folding Kinetics in a Multidomain Protein with Interwoven Chain Topology.ACS Cent Sci. 2022 Jun 22;8(6):763-774. doi: 10.1021/acscentsci.2c00140. Epub 2022 May 19. ACS Cent Sci. 2022. PMID: 35756371 Free PMC article.
-
Modeling of DNA binding to the condensin hinge domain using molecular dynamics simulations guided by atomic force microscopy.PLoS Comput Biol. 2021 Jul 30;17(7):e1009265. doi: 10.1371/journal.pcbi.1009265. eCollection 2021 Jul. PLoS Comput Biol. 2021. PMID: 34329301 Free PMC article.
-
Enhanced Conformational Sampling with an Adaptive Coarse-Grained Elastic Network Model Using Short-Time All-Atom Molecular Dynamics.J Chem Theory Comput. 2022 Apr 12;18(4):2062-2074. doi: 10.1021/acs.jctc.1c01074. Epub 2022 Mar 24. J Chem Theory Comput. 2022. PMID: 35325529 Free PMC article.
References
-
- Fersht AR. From the first protein structures to our current knowledge of protein folding: Delights and scepticisms. Nat Rev Mol Cell Biol. 2008;9:650–654. - PubMed
-
- Matouschek A, Kellis JT, Jr, Serrano L, Fersht AR. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–126. - PubMed
-
- Eaton WA, Munoz V, Thompson PA, Chan CK, Hofrichter J. Submillisecond kinetics of protein folding. Curr Opin Struct Biol. 1997;7:10–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

