Motif-optimized subtype A HIV envelope-based DNA vaccines rapidly elicit neutralizing antibodies when delivered sequentially
- PMID: 22749601
- PMCID: PMC3447634
- DOI: 10.1016/j.vaccine.2012.06.042
Motif-optimized subtype A HIV envelope-based DNA vaccines rapidly elicit neutralizing antibodies when delivered sequentially
Abstract
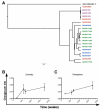
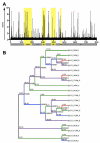
HIV-1 infection results in the development of a diverging quasispecies unique to each infected individual. Envelope (Env)-specific neutralizing antibodies (NAbs) typically develop over months to years after infection and initially are limited to the infecting virus. In some subjects, antibody responses develop that neutralize heterologous isolates (HNAbs), a phenomenon termed broadening of the NAb response. Studies of co-crystalized antibodies and proteins have facilitated the identification of some targets of broadly neutralizing monoclonal antibodies (NmAbs) capable of neutralizing many or most heterologous viruses; however, the ontogeny of these antibodies in vivo remains elusive. We hypothesize that Env protein escape variants stimulate broad NAb development in vivo and could generate such NAbs when used as immunogens. Here we test this hypothesis in rabbits using HIV Env vaccines featuring: (1) use of individual quasispecies env variants derived from an HIV-1 subtype A-infected subject exhibiting high levels of NAbs within the first year of infection that increased and broadened with time; (2) motif optimization of envs to enhance in vivo expression of DNA formulated as vaccines; and (3) a combined DNA plus protein boosting regimen. Vaccines consisted of multiple env variants delivered sequentially and a simpler regimen that utilized only the least and most divergent clones. The simpler regimen was as effective as the more complex approach in generating modest HNAbs and was more efficient when modified, motif-optimized DNA was used in combination with trimeric gp140 protein. This is a rationally designed strategy that facilitates future vaccine design by addressing the difficult problem of generating HNAbs to HIV by empirically testing the immunogenicity of naturally occurring quasispecies env variants.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Rapid Induction of Multifunctional Antibodies in Rabbits and Macaques by Clade C HIV-1 CAP257 Envelopes Circulating During Epitope-Specific Neutralization Breadth Development.Front Immunol. 2020 Jun 2;11:984. doi: 10.3389/fimmu.2020.00984. eCollection 2020. Front Immunol. 2020. PMID: 32582155 Free PMC article.
-
Envelope variants circulating as initial neutralization breadth developed in two HIV-infected subjects stimulate multiclade neutralizing antibodies in rabbits.J Virol. 2014 Nov;88(22):12949-67. doi: 10.1128/JVI.01812-14. Epub 2014 Sep 10. J Virol. 2014. PMID: 25210191 Free PMC article.
-
Prime-Boost Immunizations with DNA, Modified Vaccinia Virus Ankara, and Protein-Based Vaccines Elicit Robust HIV-1 Tier 2 Neutralizing Antibodies against the CAP256 Superinfecting Virus.J Virol. 2019 Apr 3;93(8):e02155-18. doi: 10.1128/JVI.02155-18. Print 2019 Apr 15. J Virol. 2019. PMID: 30760570 Free PMC article.
-
Passive immunization as tool to identify protective HIV-1 Env epitopes.Curr HIV Res. 2007 Nov;5(6):642-55. doi: 10.2174/157016207782418506. Curr HIV Res. 2007. PMID: 18045119 Review.
-
Neutralizing antibodies and control of HIV: moves and countermoves.Curr HIV/AIDS Rep. 2012 Mar;9(1):64-72. doi: 10.1007/s11904-011-0105-5. Curr HIV/AIDS Rep. 2012. PMID: 22203469 Review.
Cited by
-
Regulation of HIV-Gag expression and targeting to the endolysosomal/secretory pathway by the luminal domain of lysosomal-associated membrane protein (LAMP-1) enhance Gag-specific immune response.PLoS One. 2014 Jun 16;9(6):e99887. doi: 10.1371/journal.pone.0099887. eCollection 2014. PLoS One. 2014. PMID: 24932692 Free PMC article.
-
Rapid Induction of Multifunctional Antibodies in Rabbits and Macaques by Clade C HIV-1 CAP257 Envelopes Circulating During Epitope-Specific Neutralization Breadth Development.Front Immunol. 2020 Jun 2;11:984. doi: 10.3389/fimmu.2020.00984. eCollection 2020. Front Immunol. 2020. PMID: 32582155 Free PMC article.
-
HIV-1 escapes from N332-directed antibody neutralization in an elite neutralizer by envelope glycoprotein elongation and introduction of unusual disulfide bonds.Retrovirology. 2016 Jul 7;13(1):48. doi: 10.1186/s12977-016-0279-4. Retrovirology. 2016. PMID: 27388013 Free PMC article.
-
Improvement of antibody responses by HIV envelope DNA and protein co-immunization.Vaccine. 2014 Jan 16;32(4):507-13. doi: 10.1016/j.vaccine.2013.11.022. Epub 2013 Nov 23. Vaccine. 2014. PMID: 24280279 Free PMC article.
-
Tuning environmental timescales to evolve and maintain generalists.Proc Natl Acad Sci U S A. 2020 Jun 9;117(23):12693-12699. doi: 10.1073/pnas.1914586117. Epub 2020 May 26. Proc Natl Acad Sci U S A. 2020. PMID: 32457160 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

