Designing redox potential-controlled protein switches based on mutually exclusive proteins
- PMID: 22733630
- PMCID: PMC3537242
- DOI: 10.1002/pro.2109
Designing redox potential-controlled protein switches based on mutually exclusive proteins
Abstract
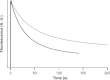
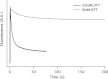
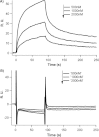
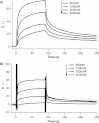
Synthetic/artificial protein switches provide an efficient means of controlling protein functions using chemical signals and stimuli. Mutually exclusive proteins, in which only the host or guest domain can remain folded at a given time owing to conformational strain, have been used to engineer novel protein switches that can switch enzymatic functions on and off in response to ligand binding. To further explore the potential of mutually exclusive proteins as protein switches and sensors, we report here a new redox-based approach to engineer a mutually exclusive folding-based protein switch. By introducing a disulfide bond into the host domain of a mutually exclusive protein, we demonstrate that it is feasible to use redox potential to switch the host domain between its folded and unfolded conformations via the mutually exclusive folding mechanism, and thus switching the functionality of the host domain on and off. Our study opens a new and potentially general avenue that uses mutually exclusive proteins to design novel switches able to control the function of a variety of proteins.
Copyright © 2012 The Protein Society.
Figures


Similar articles
-
Influence of Secondary-Structure Folding on the Mutually Exclusive Folding Process of GL5/I27 Protein: Evidence from Molecular Dynamics Simulations.Int J Mol Sci. 2016 Nov 23;17(11):1962. doi: 10.3390/ijms17111962. Int J Mol Sci. 2016. PMID: 27886109 Free PMC article.
-
Direct observation of tug-of-war during the folding of a mutually exclusive protein.J Am Chem Soc. 2009 Sep 23;131(37):13347-54. doi: 10.1021/ja903480j. J Am Chem Soc. 2009. PMID: 19719116
-
Molecular simulations of mutually exclusive folding in a two-domain protein switch.Biophys J. 2011 Feb 2;100(3):756-764. doi: 10.1016/j.bpj.2010.12.3710. Biophys J. 2011. PMID: 21281591 Free PMC article.
-
Rational design of FRET sensor proteins based on mutually exclusive domain interactions.Biochem Soc Trans. 2013 Oct;41(5):1201-5. doi: 10.1042/BST20130128. Biochem Soc Trans. 2013. PMID: 24059509 Free PMC article. Review.
-
Thiol-Disulfide Exchange Reactions in the Mammalian Extracellular Environment.Annu Rev Chem Biomol Eng. 2016 Jun 7;7:197-222. doi: 10.1146/annurev-chembioeng-080615-033553. Epub 2016 Mar 17. Annu Rev Chem Biomol Eng. 2016. PMID: 27023663 Free PMC article. Review.
Cited by
-
Rational design of a fusion protein to exhibit disulfide-mediated logic gate behavior.ACS Synth Biol. 2015 Apr 17;4(4):400-6. doi: 10.1021/sb500254g. Epub 2014 Aug 27. ACS Synth Biol. 2015. PMID: 25144732 Free PMC article.
-
Tuning response curves for synthetic biology.ACS Synth Biol. 2013 Oct 18;2(10):547-67. doi: 10.1021/sb4000564. Epub 2013 Sep 3. ACS Synth Biol. 2013. PMID: 23905721 Free PMC article.
-
Influence of Secondary-Structure Folding on the Mutually Exclusive Folding Process of GL5/I27 Protein: Evidence from Molecular Dynamics Simulations.Int J Mol Sci. 2016 Nov 23;17(11):1962. doi: 10.3390/ijms17111962. Int J Mol Sci. 2016. PMID: 27886109 Free PMC article.
-
Regulated unfolding of proteins in signaling.FEBS Lett. 2013 Apr 17;587(8):1081-8. doi: 10.1016/j.febslet.2013.02.024. Epub 2013 Feb 20. FEBS Lett. 2013. PMID: 23454209 Free PMC article. Review.
-
Protein nanomechanics in biological context.Biophys Rev. 2021 Aug 7;13(4):435-454. doi: 10.1007/s12551-021-00822-9. eCollection 2021 Aug. Biophys Rev. 2021. PMID: 34466164 Free PMC article. Review.
References
-
- Inman GJ. Switching TGFbeta from a tumor suppressor to a tumor promoter. Curr Opin Genet Dev. 2011;21:93–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

