Gut immune maturation depends on colonization with a host-specific microbiota
- PMID: 22726443
- PMCID: PMC3442780
- DOI: 10.1016/j.cell.2012.04.037
Gut immune maturation depends on colonization with a host-specific microbiota
Abstract
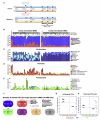
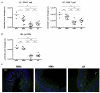
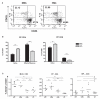
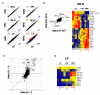
Gut microbial induction of host immune maturation exemplifies host-microbe mutualism. We colonized germ-free (GF) mice with mouse microbiota (MMb) or human microbiota (HMb) to determine whether small intestinal immune maturation depends on a coevolved host-specific microbiota. Gut bacterial numbers and phylum abundance were similar in MMb and HMb mice, but bacterial species differed, especially the Firmicutes. HMb mouse intestines had low levels of CD4(+) and CD8(+) T cells, few proliferating T cells, few dendritic cells, and low antimicrobial peptide expression--all characteristics of GF mice. Rat microbiota also failed to fully expand intestinal T cell numbers in mice. Colonizing GF or HMb mice with mouse-segmented filamentous bacteria (SFB) partially restored T cell numbers, suggesting that SFB and other MMb organisms are required for full immune maturation in mice. Importantly, MMb conferred better protection against Salmonella infection than HMb. A host-specific microbiota appears to be critical for a healthy immune system.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Host and microbes date exclusively.Cell. 2012 Jun 22;149(7):1428-30. doi: 10.1016/j.cell.2012.06.005. Cell. 2012. PMID: 22726431
-
Mucosal immunology: Any old bugs won't do.Nat Rev Immunol. 2012 Jul 25;12(8):554-5. doi: 10.1038/nri3270. Nat Rev Immunol. 2012. PMID: 22828909 No abstract available.
Similar articles
-
Growth and host interaction of mouse segmented filamentous bacteria in vitro.Nature. 2015 Apr 2;520(7545):99-103. doi: 10.1038/nature14027. Epub 2015 Jan 19. Nature. 2015. PMID: 25600271 Free PMC article.
-
Gut microbiota and lipopolysaccharide content of the diet influence development of regulatory T cells: studies in germ-free mice.BMC Immunol. 2008 Nov 6;9:65. doi: 10.1186/1471-2172-9-65. BMC Immunol. 2008. PMID: 18990206 Free PMC article.
-
Adaptive immune education by gut microbiota antigens.Immunology. 2018 May;154(1):28-37. doi: 10.1111/imm.12896. Epub 2018 Feb 8. Immunology. 2018. PMID: 29338074 Free PMC article. Review.
-
Specific Commensal Bacterium Critically Regulates Gut Microbiota Osteoimmunomodulatory Actions During Normal Postpubertal Skeletal Growth and Maturation.JBMR Plus. 2020 Jan 30;4(3):e10338. doi: 10.1002/jbm4.10338. eCollection 2020 Mar. JBMR Plus. 2020. PMID: 32161843 Free PMC article.
-
Interactions between the microbiota and the immune system.Science. 2012 Jun 8;336(6086):1268-73. doi: 10.1126/science.1223490. Epub 2012 Jun 6. Science. 2012. PMID: 22674334 Free PMC article. Review.
Cited by
-
Voided Urinary Microbiota Is Stable Over Time but Impacted by Post Void Storage.Front Cell Infect Microbiol. 2020 Aug 25;10:435. doi: 10.3389/fcimb.2020.00435. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32984068 Free PMC article.
-
The Microbiota Contributes to CD8+ T Cell Activation and Nutrient Malabsorption following Intestinal Infection with Giardia duodenalis.Infect Immun. 2016 Sep 19;84(10):2853-60. doi: 10.1128/IAI.00348-16. Print 2016 Oct. Infect Immun. 2016. PMID: 27456829 Free PMC article.
-
Immunology. Welcome to the microgenderome.Science. 2013 Mar 1;339(6123):1044-5. doi: 10.1126/science.1236226. Science. 2013. PMID: 23449586 Free PMC article.
-
Commensal bacteria and MAMPs are necessary for stress-induced increases in IL-1β and IL-18 but not IL-6, IL-10 or MCP-1.PLoS One. 2012;7(12):e50636. doi: 10.1371/journal.pone.0050636. Epub 2012 Dec 7. PLoS One. 2012. PMID: 23236381 Free PMC article.
-
The gut microbiota and inflammatory bowel diseases.Transl Res. 2017 Jan;179:38-48. doi: 10.1016/j.trsl.2016.06.002. Epub 2016 Jun 14. Transl Res. 2017. PMID: 27371886 Free PMC article. Review.
References
-
- Chassin C, Kocur M, Pott J, Duerr CU, Gütle D, Lotz M, Hornef MW. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. 2010;8:358–368. - PubMed
-
- Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr. Opin. Immunol. 2010;22:455–460. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

