Fitness analyses of all possible point mutations for regions of genes in yeast
- PMID: 22722372
- PMCID: PMC3509169
- DOI: 10.1038/nprot.2012.069
Fitness analyses of all possible point mutations for regions of genes in yeast
Abstract
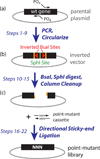
Deep sequencing can accurately measure the relative abundance of hundreds of mutations in a single bulk competition experiment, which can give a direct readout of the fitness of each mutant. Here we describe a protocol that we previously developed and optimized to measure the fitness effects of all possible individual codon substitutions for 10-aa regions of essential genes in yeast. Starting with a conditional strain (i.e., a temperature-sensitive strain), we describe how to efficiently generate plasmid libraries of point mutants that can then be transformed to generate libraries of yeast. The yeast libraries are competed under conditions that select for mutant function. Deep-sequencing analyses are used to determine the relative fitness of all mutants. This approach is faster and cheaper per mutant compared with analyzing individually isolated mutants. The protocol can be performed in ∼4 weeks and many 10-aa regions can be analyzed in parallel.
Figures



Similar articles
-
Defining essential genes and identifying virulence factors of Porphyromonas gingivalis by massively parallel sequencing of transposon libraries (Tn-seq).Methods Mol Biol. 2015;1279:25-43. doi: 10.1007/978-1-4939-2398-4_3. Methods Mol Biol. 2015. PMID: 25636611 Free PMC article.
-
Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons.mBio. 2015 May 12;6(3):e00306-15. doi: 10.1128/mBio.00306-15. mBio. 2015. PMID: 25968644 Free PMC article.
-
Experimental illumination of a fitness landscape.Proc Natl Acad Sci U S A. 2011 May 10;108(19):7896-901. doi: 10.1073/pnas.1016024108. Epub 2011 Apr 4. Proc Natl Acad Sci U S A. 2011. PMID: 21464309 Free PMC article.
-
Residue specific contributions to stability and activity inferred from saturation mutagenesis and deep sequencing.Curr Opin Struct Biol. 2014 Feb;24:63-71. doi: 10.1016/j.sbi.2013.12.001. Epub 2014 Jan 7. Curr Opin Struct Biol. 2014. PMID: 24721454 Review.
-
Viewing protein fitness landscapes through a next-gen lens.Genetics. 2014 Oct;198(2):461-71. doi: 10.1534/genetics.114.168351. Genetics. 2014. PMID: 25316787 Free PMC article. Review.
Cited by
-
Engineered control of enzyme structural dynamics and function.Protein Sci. 2018 Apr;27(4):825-838. doi: 10.1002/pro.3379. Epub 2018 Feb 16. Protein Sci. 2018. PMID: 29380452 Free PMC article. Review.
-
Correlation of fitness landscapes from three orthologous TIM barrels originates from sequence and structure constraints.Nat Commun. 2017 Mar 6;8:14614. doi: 10.1038/ncomms14614. Nat Commun. 2017. PMID: 28262665 Free PMC article.
-
Mutational fitness landscape and drug resistance.Curr Opin Struct Biol. 2023 Feb;78:102525. doi: 10.1016/j.sbi.2022.102525. Epub 2023 Jan 6. Curr Opin Struct Biol. 2023. PMID: 36621152 Free PMC article. Review.
-
Deep mutational scanning: A versatile tool in systematically mapping genotypes to phenotypes.Front Genet. 2023 Jan 12;14:1087267. doi: 10.3389/fgene.2023.1087267. eCollection 2023. Front Genet. 2023. PMID: 36713072 Free PMC article. Review.
-
Inferring the distribution of fitness effects in patient-sampled and experimental virus populations: two case studies.Heredity (Edinb). 2022 Feb;128(2):79-87. doi: 10.1038/s41437-021-00493-y. Epub 2022 Jan 5. Heredity (Edinb). 2022. PMID: 34987185 Free PMC article.
References
-
- Jensen JD, Wong A, Aquadro CF. Approaches for identifying targets of positive selection. Trends Genet. 2007;23:568–577. - PubMed
-
- Hegreness M, Shoresh N, Hartl D, Kishony R. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science. 2006;311:1615–1617. - PubMed
-
- Lind PA, Berg OG, Andersson DI. Mutational robustness of ribosomal protein genes. Science. 2010;330:825–827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

