Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer
- PMID: 22706305
- PMCID: PMC3386824
- DOI: 10.1172/JCI62613
Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer
Abstract
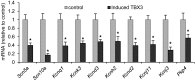
The contraction pattern of the heart relies on the activation and conduction of the electrical impulse. Perturbations of cardiac conduction have been associated with congenital and acquired arrhythmias as well as cardiac arrest. The pattern of conduction depends on the regulation of heterogeneous gene expression by key transcription factors and transcriptional enhancers. Here, we assessed the genome-wide occupation of conduction system-regulating transcription factors TBX3, NKX2-5, and GATA4 and of enhancer-associated coactivator p300 in the mouse heart, uncovering cardiac enhancers throughout the genome. Many of the enhancers colocalized with ion channel genes repressed by TBX3, including the clustered sodium channel genes Scn5a, essential for cardiac function, and Scn10a. We identified 2 enhancers in the Scn5a/Scn10a locus, which were regulated by TBX3 and its family member and activator, TBX5, and are functionally conserved in humans. We also provided evidence that a SNP in the SCN10A enhancer associated with alterations in cardiac conduction patterns in humans disrupts TBX3/TBX5 binding and reduces the cardiac activity of the enhancer in vivo. Thus, the identification of key regulatory elements for cardiac conduction helps to explain how genetic variants in noncoding regulatory DNA sequences influence the regulation of cardiac conduction and the predisposition for cardiac arrhythmias.
Figures



Similar articles
-
Variant Intronic Enhancer Controls SCN10A-short Expression and Heart Conduction.Circulation. 2021 Jul 20;144(3):229-242. doi: 10.1161/CIRCULATIONAHA.121.054083. Epub 2021 Apr 29. Circulation. 2021. PMID: 33910361
-
A common genetic variant within SCN10A modulates cardiac SCN5A expression.J Clin Invest. 2014 Apr;124(4):1844-52. doi: 10.1172/JCI73140. Epub 2014 Mar 18. J Clin Invest. 2014. PMID: 24642470 Free PMC article.
-
TBX5 drives Scn5a expression to regulate cardiac conduction system function.J Clin Invest. 2012 Jul;122(7):2509-18. doi: 10.1172/JCI62617. Epub 2012 Jun 25. J Clin Invest. 2012. PMID: 22728936 Free PMC article.
-
From GWAS to function: genetic variation in sodium channel gene enhancer influences electrical patterning.Trends Cardiovasc Med. 2014 Apr;24(3):99-104. doi: 10.1016/j.tcm.2013.09.001. Epub 2013 Dec 17. Trends Cardiovasc Med. 2014. PMID: 24360055 Review.
-
Gene regulatory elements of the cardiac conduction system.Brief Funct Genomics. 2014 Jan;13(1):28-38. doi: 10.1093/bfgp/elt031. Epub 2013 Aug 22. Brief Funct Genomics. 2014. PMID: 23969024 Review.
Cited by
-
Role of Genetic Variation in Transcriptional Regulatory Elements in Heart Rhythm.Cells. 2023 Dec 19;13(1):4. doi: 10.3390/cells13010004. Cells. 2023. PMID: 38201209 Free PMC article. Review.
-
SNPs identified as modulators of ECG traits in the general population do not markedly affect ECG traits during acute myocardial infarction nor ventricular fibrillation risk in this condition.PLoS One. 2013;8(2):e57216. doi: 10.1371/journal.pone.0057216. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437344 Free PMC article.
-
Association of common and rare variants of SCN10A gene with sudden unexplained nocturnal death syndrome in Chinese Han population.Int J Legal Med. 2017 Jan;131(1):53-60. doi: 10.1007/s00414-016-1397-1. Epub 2016 Jun 7. Int J Legal Med. 2017. PMID: 27272739
-
Spatiotemporal regulation of enhancers during cardiogenesis.Cell Mol Life Sci. 2017 Jan;74(2):257-265. doi: 10.1007/s00018-016-2322-y. Epub 2016 Aug 6. Cell Mol Life Sci. 2017. PMID: 27497925 Free PMC article. Review.
-
An enhancer cluster controls gene activity and topology of the SCN5A-SCN10A locus in vivo.Nat Commun. 2019 Oct 30;10(1):4943. doi: 10.1038/s41467-019-12856-5. Nat Commun. 2019. PMID: 31666509 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

