Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications
- PMID: 22704344
- PMCID: PMC3365388
- DOI: 10.1007/s13148-011-0032-8
Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications
Abstract
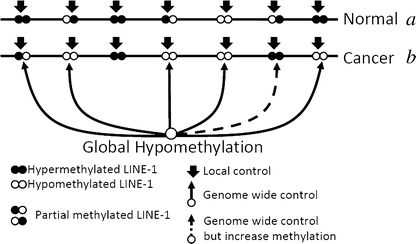
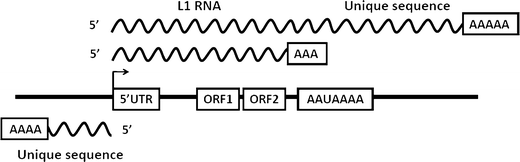
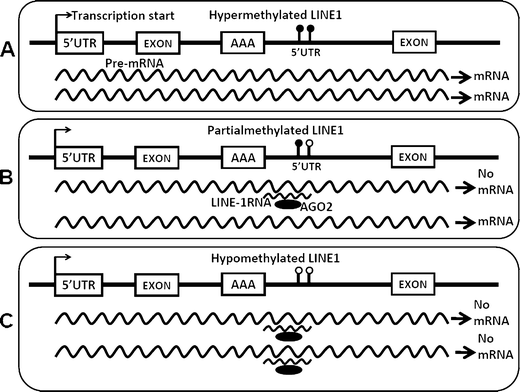
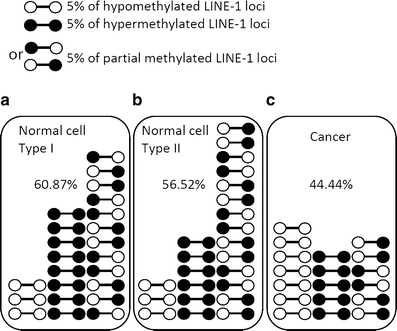
Epigenetic changes in long interspersed nuclear element-1s (LINE-1s or L1s) occur early during the process of carcinogenesis. A lower methylation level (hypomethylation) of LINE-1 is common in most cancers, and the methylation level is further decreased in more advanced cancers. Consequently, several previous studies have suggested the use of LINE-1 hypomethylation levels in cancer screening, risk assessment, tumor staging, and prognostic prediction. Epigenomic changes are complex, and global hypomethylation influences LINE-1s in a generalized fashion. However, the methylation levels of some loci are dependent on their locations. The consequences of LINE-1 hypomethylation are genomic instability and alteration of gene expression. There are several mechanisms that promote both of these consequences in cis. Therefore, the methylation levels of different sets of LINE-1s may represent certain phenotypes. Furthermore, the methylation levels of specific sets of LINE-1s may indicate carcinogenesis-dependent hypomethylation. LINE-1 methylation pattern analysis can classify LINE-1s into one of three classes based on the number of methylated CpG dinucleotides. These classes include hypermethylation, partial methylation, and hypomethylation. The number of partial and hypermethylated loci, but not hypomethylated LINE-1s, is different among normal cell types. Consequently, the number of hypomethylated loci is a more promising marker than methylation level in the detection of cancer DNA. Further genome-wide studies to measure the methylation level of each LINE-1 locus may improve PCR-based methylation analysis to allow for a more specific and sensitive detection of cancer DNA or for an analysis of certain cancer phenotypes.
Electronic supplementary material: The online version of this article (doi:10.1007/s13148-011-0032-8) contains supplementary material, which is available to authorized users.
Figures
Similar articles
-
Patterns and possible roles of LINE-1 methylation changes in smoke-exposed epithelia.PLoS One. 2012;7(9):e45292. doi: 10.1371/journal.pone.0045292. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028911 Free PMC article.
-
Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2.PLoS One. 2011 Mar 15;6(3):e17934. doi: 10.1371/journal.pone.0017934. PLoS One. 2011. PMID: 21423624 Free PMC article.
-
LINE-1 methylation patterns of different loci in normal and cancerous cells.Nucleic Acids Res. 2008 Oct;36(17):5704-12. doi: 10.1093/nar/gkn571. Epub 2008 Sep 6. Nucleic Acids Res. 2008. PMID: 18776216 Free PMC article.
-
Long Interspersed Element-1 Methylation Level as a Prognostic Biomarker in Gastrointestinal Cancers.Digestion. 2018;97(1):26-30. doi: 10.1159/000484104. Epub 2018 Feb 1. Digestion. 2018. PMID: 29393154 Review.
-
Cancer-linked DNA hypomethylation and its relationship to hypermethylation.Curr Top Microbiol Immunol. 2006;310:251-74. doi: 10.1007/3-540-31181-5_12. Curr Top Microbiol Immunol. 2006. PMID: 16909914 Review.
Cited by
-
Patterns and possible roles of LINE-1 methylation changes in smoke-exposed epithelia.PLoS One. 2012;7(9):e45292. doi: 10.1371/journal.pone.0045292. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028911 Free PMC article.
-
Evaluation of NID2 promoter methylation for screening of Oral squamous cell carcinoma.BMC Cancer. 2020 Mar 14;20(1):218. doi: 10.1186/s12885-020-6692-z. BMC Cancer. 2020. PMID: 32171289 Free PMC article.
-
Epigenetic regulation of cancer-associated fibroblast heterogeneity.Biochim Biophys Acta Rev Cancer. 2023 May;1878(3):188901. doi: 10.1016/j.bbcan.2023.188901. Epub 2023 Apr 28. Biochim Biophys Acta Rev Cancer. 2023. PMID: 37120098 Free PMC article. Review.
-
Tumor Long Interspersed Nucleotide Element-1 (LINE-1) Hypomethylation in Relation to Age of Colorectal Cancer Diagnosis and Prognosis.Cancers (Basel). 2021 Apr 22;13(9):2016. doi: 10.3390/cancers13092016. Cancers (Basel). 2021. PMID: 33922024 Free PMC article.
-
Clinical advances in molecular biomarkers for cancer diagnosis and therapy.Int J Mol Sci. 2013 Jul 16;14(7):14771-84. doi: 10.3390/ijms140714771. Int J Mol Sci. 2013. PMID: 23863689 Free PMC article. Review.
References
-
- Ahn JB, Chung WB, Maeda O, Shin SJ, Kim HS, Chung HC, Kim NK, Issa JP (2011) DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. doi:10.1002/cncr.25737 - DOI - PMC - PubMed
-
- An B, Kondo Y, Okamoto Y, Shinjo K, Kanemitsu Y, Komori K, Hirai T, Sawaki A, Tajika M, Nakamura T, Yamao K, Yatabe Y, Fujii M, Murakami H, Osada H, Tani T, Matsuo K, Shen L, Issa JP, Sekido Y. Characteristic methylation profile in CpG island methylator phenotype-negative distal colorectal cancers. Int J Cancer. 2010;127(9):2095–2105. doi: 10.1002/ijc.25225. - DOI - PubMed
-
- Aparicio A, North B, Barske L, Wang X, Bollati V, Weisenberger D, Yoo C, Tannir N, Horne E, Groshen S, Jones P, Yang A, Issa JP. LINE-1 methylation in plasma DNA as a biomarker of activity of DNA methylation inhibitors in patients with solid tumors. Epigenetics. 2009;4(3):176–184. doi: 10.4161/epi.4.3.8694. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
