RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis
- PMID: 22689894
- PMCID: PMC3425185
- DOI: 10.1104/pp.112.199646
RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis
Abstract
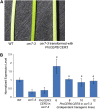
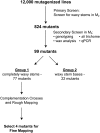
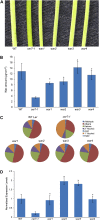
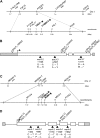
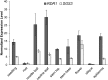
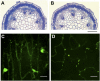
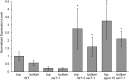
The cuticle is a protective layer that coats the primary aerial surfaces of land plants and mediates plant interactions with the environment. It is synthesized by epidermal cells and is composed of a cutin polyester matrix that is embedded and covered with cuticular waxes. Recently, we have discovered a novel regulatory mechanism of cuticular wax biosynthesis that involves the ECERIFERUM7 (CER7) ribonuclease, a core subunit of the exosome. We hypothesized that at the onset of wax production, the CER7 ribonuclease degrades an mRNA specifying a repressor of CER3, a wax biosynthetic gene whose protein product is required for wax formation via the decarbonylation pathway. In the absence of this repressor, CER3 is expressed, leading to wax production. To identify the putative repressor of CER3 and to unravel the mechanism of CER7-mediated regulation of wax production, we performed a screen for suppressors of the cer7 mutant. Our screen resulted in the isolation of components of the RNA-silencing machinery, RNA-DEPENDENT RNA POLYMERASE1 and SUPPRESSOR OF GENE SILENCING3, implicating RNA silencing in the control of cuticular wax deposition during inflorescence stem development in Arabidopsis (Arabidopsis thaliana).
Figures

Similar articles
-
The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development.Plant Physiol. 2015 Feb;167(2):323-36. doi: 10.1104/pp.114.252825. Epub 2014 Dec 11. Plant Physiol. 2015. PMID: 25502190 Free PMC article.
-
SUPERKILLER Complex Components Are Required for the RNA Exosome-Mediated Control of Cuticular Wax Biosynthesis in Arabidopsis Inflorescence Stems.Plant Physiol. 2016 Jun;171(2):960-73. doi: 10.1104/pp.16.00450. Epub 2016 Apr 28. Plant Physiol. 2016. PMID: 27208312 Free PMC article.
-
A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis.Plant Cell. 2007 Mar;19(3):904-13. doi: 10.1105/tpc.106.049304. Epub 2007 Mar 9. Plant Cell. 2007. PMID: 17351114 Free PMC article.
-
Regulatory mechanisms underlying cuticular wax biosynthesis.J Exp Bot. 2022 May 13;73(9):2799-2816. doi: 10.1093/jxb/erab509. J Exp Bot. 2022. PMID: 35560199 Review.
-
Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species.Plant Cell Rep. 2015 Apr;34(4):557-72. doi: 10.1007/s00299-015-1772-2. Epub 2015 Feb 19. Plant Cell Rep. 2015. PMID: 25693495 Review.
Cited by
-
Differential expression of miRNAs and their targets in wax-deficient rapeseed.Sci Rep. 2019 Aug 21;9(1):12201. doi: 10.1038/s41598-019-48439-z. Sci Rep. 2019. PMID: 31434948 Free PMC article.
-
CsCER6 and CsCER7 Influence Fruit Glossiness by Regulating Fruit Cuticular Wax Accumulation in Cucumber.Int J Mol Sci. 2023 Jan 6;24(2):1135. doi: 10.3390/ijms24021135. Int J Mol Sci. 2023. PMID: 36674649 Free PMC article.
-
Analysis of the RNA-Dependent RNA Polymerase 1 (RDR1) Gene Family in Melon.Plants (Basel). 2022 Jul 7;11(14):1795. doi: 10.3390/plants11141795. Plants (Basel). 2022. PMID: 35890429 Free PMC article.
-
Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis.Planta. 2019 May;249(5):1627-1643. doi: 10.1007/s00425-019-03115-4. Epub 2019 Mar 2. Planta. 2019. PMID: 30826884
-
AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types.Plant J. 2014 Oct;80(2):216-29. doi: 10.1111/tpj.12624. Epub 2014 Aug 26. Plant J. 2014. PMID: 25060192 Free PMC article.
References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Barthlott W, Neinhuis C. (1997) Purity of the sacred lotus, or escape from contamination in biological surfaces. Planta 202: 1–8
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

