Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase
- PMID: 22689655
- PMCID: PMC3373407
- DOI: 10.1083/jcb.201203061
Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase
Abstract
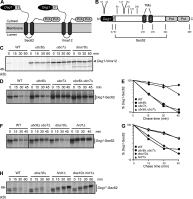
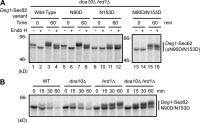
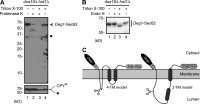
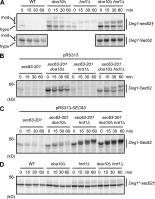
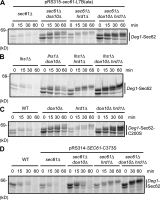
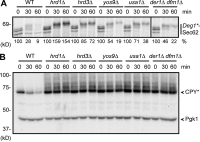
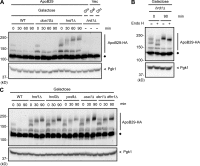
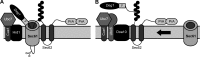
Little is known about quality control of proteins that aberrantly or persistently engage the endoplasmic reticulum (ER)-localized translocon en route to membrane localization or the secretory pathway. Hrd1 and Doa10, the primary ubiquitin ligases that function in ER-associated degradation (ERAD) in yeast, target distinct subsets of misfolded or otherwise abnormal proteins based primarily on degradation signal (degron) location. We report the surprising observation that fusing Deg1, a cytoplasmic degron normally recognized by Doa10, to the Sec62 membrane protein rendered the protein a Hrd1 substrate. Hrd1-dependent degradation occurred when Deg1-Sec62 aberrantly engaged the Sec61 translocon channel and underwent topological rearrangement. Mutations that prevent translocon engagement caused a reversion to Doa10-dependent degradation. Similarly, a variant of apolipoprotein B, a protein known to be cotranslocationally targeted for proteasomal degradation, was also a Hrd1 substrate. Hrd1 therefore likely plays a general role in targeting proteins that persistently associate with and potentially obstruct the translocon.
Figures
Similar articles
-
Endoplasmic reticulum stress differentially inhibits endoplasmic reticulum and inner nuclear membrane protein quality control degradation pathways.J Biol Chem. 2019 Dec 20;294(51):19814-19830. doi: 10.1074/jbc.RA119.010295. Epub 2019 Nov 13. J Biol Chem. 2019. PMID: 31723032 Free PMC article.
-
Overlapping function of Hrd1 and Ste24 in translocon quality control provides robust channel surveillance.J Biol Chem. 2020 Nov 20;295(47):16113-16120. doi: 10.1074/jbc.AC120.016191. Epub 2020 Oct 8. J Biol Chem. 2020. PMID: 33033070 Free PMC article.
-
N-terminal acetylation of the yeast Derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates.Mol Biol Cell. 2013 Apr;24(7):890-900. doi: 10.1091/mbc.E12-11-0838. Epub 2013 Jan 30. Mol Biol Cell. 2013. PMID: 23363603 Free PMC article.
-
[Physiological Roles of Ubiquitin Ligases Related to the Endoplasmic Reticulum].Yakugaku Zasshi. 2016;136(6):805-9. doi: 10.1248/yakushi.15-00292-2. Yakugaku Zasshi. 2016. PMID: 27252059 Review. Japanese.
-
Proteolytic regulation of metabolic enzymes by E3 ubiquitin ligase complexes: lessons from yeast.Crit Rev Biochem Mol Biol. 2015;50(6):489-502. doi: 10.3109/10409238.2015.1081869. Epub 2015 Sep 11. Crit Rev Biochem Mol Biol. 2015. PMID: 26362128 Review.
Cited by
-
Glucose concentration does not affect degradation of a protein that aberrantly engages the endoplasmic reticulum translocon.MicroPubl Biol. 2020;2020:248. Epub 2020 May 8. MicroPubl Biol. 2020. PMID: 32548573 Free PMC article. No abstract available.
-
Characterization of endoplasmic reticulum-associated degradation in the human fungal pathogen Candida albicans.PeerJ. 2023 Aug 25;11:e15897. doi: 10.7717/peerj.15897. eCollection 2023. PeerJ. 2023. PMID: 37645016 Free PMC article.
-
Rkr1/Ltn1 Ubiquitin Ligase-mediated Degradation of Translationally Stalled Endoplasmic Reticulum Proteins.J Biol Chem. 2015 Jul 24;290(30):18454-66. doi: 10.1074/jbc.M115.663559. Epub 2015 Jun 8. J Biol Chem. 2015. PMID: 26055716 Free PMC article.
-
Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope.Crit Rev Biochem Mol Biol. 2015 Jan-Feb;50(1):1-17. doi: 10.3109/10409238.2014.959889. Epub 2014 Sep 18. Crit Rev Biochem Mol Biol. 2015. PMID: 25231236 Free PMC article. Review.
-
Three old and one new: protein import into red algal-derived plastids surrounded by four membranes.Protoplasma. 2013 Oct;250(5):1013-23. doi: 10.1007/s00709-013-0498-7. Epub 2013 Apr 24. Protoplasma. 2013. PMID: 23612938 Review.
References
-
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. 1989. Current protocols in molecular biology. Wiley, New York
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

