Osteopetrosis mutation R444L causes endoplasmic reticulum retention and misprocessing of vacuolar H+-ATPase a3 subunit
- PMID: 22685294
- PMCID: PMC3411020
- DOI: 10.1074/jbc.M112.345702
Osteopetrosis mutation R444L causes endoplasmic reticulum retention and misprocessing of vacuolar H+-ATPase a3 subunit
Abstract
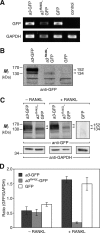
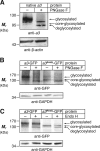
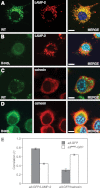
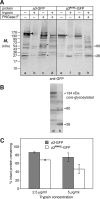
Osteopetrosis is a genetic bone disease characterized by increased bone density and fragility. The R444L missense mutation in the human V-ATPase a3 subunit (TCIRG1) is one of several known mutations in a3 and other proteins that can cause this disease. The autosomal recessive R444L mutation results in a particularly malignant form of infantile osteopetrosis that is lethal in infancy, or early childhood. We have studied this mutation using the pMSCV retroviral vector system to integrate the cDNA construct for green fluorescent protein (GFP)-fused a3(R445L) mutant protein into the RAW 264.7 mouse osteoclast differentiation model. In comparison with wild-type a3, the mutant glycoprotein localized to the ER instead of lysosomes and its oligosaccharide moiety was misprocessed, suggesting inability of the core-glycosylated glycoprotein to traffic to the Golgi. Reduced steady-state expression of the mutant protein, in comparison with wild type, suggested that the former was being degraded, likely through the endoplasmic reticulum-associated degradation pathway. In differentiated osteoclasts, a3(R445L) was found to degrade at an increased rate over the course of osteoclastogenesis. Limited proteolysis studies suggested that the R445L mutation alters mouse a3 protein conformation. Together, these data suggest that Arg-445 plays a role in protein folding, or stability, and that infantile malignant osteopetrosis caused by the R444L mutation in the human V-ATPase a3 subunit is another member of the growing class of protein folding diseases. This may have implications for early-intervention treatment, using protein rescue strategies.
Figures

Similar articles
-
Identification and in silico characterization of a novel p.P208PfsX1 mutation in V-ATPase a3 subunit associated with autosomal recessive osteopetrosis in a Pakistani family.BMC Med Genet. 2017 Dec 13;18(1):148. doi: 10.1186/s12881-017-0506-4. BMC Med Genet. 2017. PMID: 29237407 Free PMC article.
-
V-ATPase a3 isoform mutations identified in osteopetrosis patients abolish its expression and disrupt osteoclast function.Exp Cell Res. 2020 Apr 15;389(2):111901. doi: 10.1016/j.yexcr.2020.111901. Epub 2020 Feb 8. Exp Cell Res. 2020. PMID: 32045577
-
Novel c.G630A TCIRG1 mutation causes aberrant splicing resulting in an unusually mild form of autosomal recessive osteopetrosis.J Cell Biochem. 2019 Oct;120(10):17180-17193. doi: 10.1002/jcb.28979. Epub 2019 May 20. J Cell Biochem. 2019. PMID: 31111556
-
Human osteopetroses and the osteoclast V-H+-ATPase enzyme system.Front Biosci. 2005 Sep 1;10:2940-54. doi: 10.2741/1750. Front Biosci. 2005. PMID: 15970548 Review.
-
The V-ATPase a3 Subunit: Structure, Function and Therapeutic Potential of an Essential Biomolecule in Osteoclastic Bone Resorption.Int J Mol Sci. 2021 Jun 28;22(13):6934. doi: 10.3390/ijms22136934. Int J Mol Sci. 2021. PMID: 34203247 Free PMC article. Review.
Cited by
-
Chemical probes and drug leads from advances in synthetic planning and methodology.Nat Rev Drug Discov. 2018 May;17(5):333-352. doi: 10.1038/nrd.2018.53. Epub 2018 Apr 13. Nat Rev Drug Discov. 2018. PMID: 29651105 Free PMC article. Review.
-
Lysosomal dysfunction in proteinopathic neurodegenerative disorders: possible therapeutic roles of cAMP and zinc.Mol Brain. 2019 Mar 12;12(1):18. doi: 10.1186/s13041-019-0439-2. Mol Brain. 2019. PMID: 30866990 Free PMC article. Review.
-
Endoplasmic Reticulum-Targeted Subunit Toxins Provide a New Approach to Rescue Misfolded Mutant Proteins and Revert Cell Models of Genetic Diseases.PLoS One. 2016 Dec 9;11(12):e0166948. doi: 10.1371/journal.pone.0166948. eCollection 2016. PLoS One. 2016. PMID: 27935997 Free PMC article.
-
Discovery of a Small-Molecule Probe for V-ATPase Function.J Am Chem Soc. 2015 Apr 29;137(16):5563-8. doi: 10.1021/jacs.5b02150. Epub 2015 Apr 20. J Am Chem Soc. 2015. PMID: 25860544 Free PMC article.
-
Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease.Ageing Res Rev. 2016 Dec;32:75-88. doi: 10.1016/j.arr.2016.05.004. Epub 2016 May 16. Ageing Res Rev. 2016. PMID: 27197071 Free PMC article. Review.
References
-
- Grüber G., Marshansky V. (2008) New insights into structure-function relationships between archeal ATP synthase (A1A0) and vacuolar-type ATPase (V1V0). Bioessays 30, 1096–1109 - PubMed
-
- Saroussi S., Nelson N. (2009) The little we know on the structure and machinery of V-ATPase. J. Exp. Biol. 212, 1604–1610 - PubMed
-
- Saroussi S., Nelson N. (2009) Vacuolar H+-ATPases. An enzyme for all seasons. Pfluegers Arch./Eur. J. Physiol. 457, 581–587 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

