Tyrosine kinase pathways modulate tumor susceptibility to natural killer cells
- PMID: 22684105
- PMCID: PMC3386806
- DOI: 10.1172/JCI58457
Tyrosine kinase pathways modulate tumor susceptibility to natural killer cells
Abstract
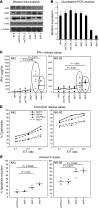
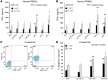
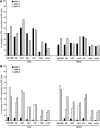
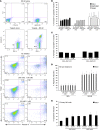
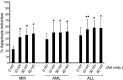
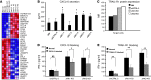
Natural killer (NK) cells are primary effectors of innate immunity directed against transformed tumor cells. In response, tumor cells have developed mechanisms to evade NK cell-mediated lysis through molecular mechanisms that are not well understood. In the present study, we used a lentiviral shRNA library targeting more than 1,000 human genes to identify 83 genes that promote target cell resistance to human NK cell-mediated killing. Many of the genes identified in this genetic screen belong to common signaling pathways; however, none of them have previously been known to modulate susceptibility of human tumor cells to immunologic destruction. Gene silencing of two members of the JAK family (JAK1 and JAK2) increased the susceptibility of a variety of tumor cell types to NK-mediated lysis and induced increased secretion of IFN-γ by NK cells. Treatment of tumor cells with JAK inhibitors also increased susceptibility to NK cell activity. These findings may have important clinical implications and suggest that small molecule inhibitors of tyrosine kinases being developed as therapeutic antitumor agents may also have significant immunologic effects in vivo.
Figures




Similar articles
-
Natural Killer Cells Suppress T Cell-Associated Tumor Immune Evasion.Cell Rep. 2019 Sep 10;28(11):2784-2794.e5. doi: 10.1016/j.celrep.2019.08.017. Cell Rep. 2019. PMID: 31509742
-
Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition.Proc Natl Acad Sci U S A. 2014 Jun 3;111(22):8025-30. doi: 10.1073/pnas.1401180111. Epub 2014 May 19. Proc Natl Acad Sci U S A. 2014. PMID: 24843152 Free PMC article.
-
JAK kinases overexpression promotes in vitro cell transformation.Oncogene. 2008 Mar 6;27(11):1511-9. doi: 10.1038/sj.onc.1210800. Epub 2007 Sep 17. Oncogene. 2008. PMID: 17873904
-
The molecular regulation of Janus kinase (JAK) activation.Biochem J. 2014 Aug 15;462(1):1-13. doi: 10.1042/BJ20140712. Biochem J. 2014. PMID: 25057888 Free PMC article. Review.
-
Inhibitors of JAK2 and JAK3: an update on the patent literature 2010 - 2012.Expert Opin Ther Pat. 2013 Apr;23(4):449-501. doi: 10.1517/13543776.2013.765862. Epub 2013 Feb 1. Expert Opin Ther Pat. 2013. PMID: 23367873 Review.
Cited by
-
c-Abl modulates tumor cell sensitivity to antibody-dependent cellular cytotoxicity.Cancer Immunol Res. 2014 Dec;2(12):1186-98. doi: 10.1158/2326-6066.CIR-14-0083. Epub 2014 Oct 9. Cancer Immunol Res. 2014. PMID: 25300860 Free PMC article.
-
Pretransplant IgG reactivity to apoptotic cells correlates with late kidney allograft loss.Am J Transplant. 2014 Jul;14(7):1581-91. doi: 10.1111/ajt.12763. Epub 2014 Jun 16. Am J Transplant. 2014. PMID: 24935695 Free PMC article.
-
A high-throughput RNAi screen for detection of immune-checkpoint molecules that mediate tumor resistance to cytotoxic T lymphocytes.EMBO Mol Med. 2015 Apr;7(4):450-63. doi: 10.15252/emmm.201404414. EMBO Mol Med. 2015. PMID: 25691366 Free PMC article.
-
Present and Future of Allogeneic Natural Killer Cell Therapy.Front Immunol. 2015 Jun 3;6:286. doi: 10.3389/fimmu.2015.00286. eCollection 2015. Front Immunol. 2015. PMID: 26089823 Free PMC article. Review.
-
A High-Throughput Assay for Screening of Natural Products that Enhanced Tumoricidal Activity of NK Cells.Biol Proced Online. 2015 Oct 28;17:12. doi: 10.1186/s12575-015-0026-6. eCollection 2015. Biol Proced Online. 2015. PMID: 26516316 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

