Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression
- PMID: 22669881
- PMCID: PMC3406894
- DOI: 10.1105/tpc.112.098749
Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression
Abstract
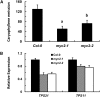
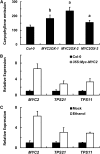
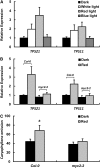
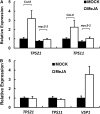
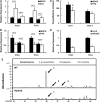
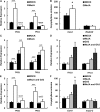
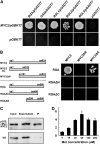
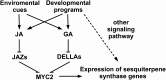
Arabidopsis thaliana flowers emit volatile terpenes, which may function in plant-insect interactions. Here, we report that Arabidopsis MYC2, a basic helix-loop-helix transcription factor, directly binds to promoters of the sesquiterpene synthase genes TPS21 and TPS11 and activates their expression. Expression of TPS21 and TPS11 can be induced by the phytohormones gibberellin (GA) and jasmonate (JA), and both inductions require MYC2. The induction of TPS21 and TPS11 results in increased emission of sesquiterpene, especially (E)-β-caryophyllene. DELLAs, the GA signaling repressors, negatively affect sesquiterpene biosynthesis, as the sesquiterpene synthase genes were repressed in plants overaccumulating REPRESSOR OF GA1-3 (RGA), one of the Arabidopsis DELLAs, and upregulated in a penta DELLA-deficient mutant. Yeast two-hybrid and coimmunoprecipitation assays demonstrated that DELLAs, represented by RGA, directly interact with MYC2. In yeast cells, the N terminus of MYC2 was responsible for binding to RGA. MYC2 has been proposed as a major mediator of JA signaling and crosstalk with abscisic acid, ethylene, and light signaling pathways. Our results demonstrate that MYC2 is also connected to GA signaling in regulating a subset of genes. In Arabidopsis inflorescences, it integrates both GA and JA signals into transcriptional regulation of sesquiterpene synthase genes and promotes sesquiterpene production.
Figures

Similar articles
-
The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses.Plant Cell. 2012 Aug;24(8):3307-19. doi: 10.1105/tpc.112.101428. Epub 2012 Aug 14. Plant Cell. 2012. PMID: 22892320 Free PMC article.
-
Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade.Proc Natl Acad Sci U S A. 2012 May 8;109(19):E1192-200. doi: 10.1073/pnas.1201616109. Epub 2012 Apr 23. Proc Natl Acad Sci U S A. 2012. PMID: 22529386 Free PMC article.
-
DELLAs modulate jasmonate signaling via competitive binding to JAZs.Dev Cell. 2010 Dec 14;19(6):884-94. doi: 10.1016/j.devcel.2010.10.024. Dev Cell. 2010. PMID: 21145503
-
JAZ repressors and the orchestration of phytohormone crosstalk.Trends Plant Sci. 2012 Jan;17(1):22-31. doi: 10.1016/j.tplants.2011.10.006. Epub 2011 Nov 21. Trends Plant Sci. 2012. PMID: 22112386 Review.
-
The plant Mediator complex and its role in jasmonate signaling.J Exp Bot. 2019 Jul 5;70(13):3415-3424. doi: 10.1093/jxb/erz233. J Exp Bot. 2019. PMID: 31089685 Free PMC article. Review.
Cited by
-
Functional Characterization of a Dendrobium officinale Geraniol Synthase DoGES1 Involved in Floral Scent Formation.Int J Mol Sci. 2020 Sep 23;21(19):7005. doi: 10.3390/ijms21197005. Int J Mol Sci. 2020. PMID: 32977586 Free PMC article.
-
Identification of long non-coding RNAs involved in floral scent of Rosa hybrida.Front Plant Sci. 2022 Oct 4;13:996474. doi: 10.3389/fpls.2022.996474. eCollection 2022. Front Plant Sci. 2022. PMID: 36267940 Free PMC article.
-
Genome-wide analysis reveals diverged patterns of codon bias, gene expression, and rates of sequence evolution in picea gene families.Genome Biol Evol. 2015 Mar 5;7(4):1002-15. doi: 10.1093/gbe/evv044. Genome Biol Evol. 2015. PMID: 25747252 Free PMC article.
-
Functional characterization of a terpene synthase responsible for (E)-β-ocimene biosynthesis identified in Pyrus betuleafolia transcriptome after herbivory.Front Plant Sci. 2022 Nov 21;13:1077229. doi: 10.3389/fpls.2022.1077229. eCollection 2022. Front Plant Sci. 2022. PMID: 36479507 Free PMC article.
-
Mediation of JA signalling in glandular trichomes by the woolly/SlMYC1 regulatory module improves pest resistance in tomato.Plant Biotechnol J. 2021 Feb;19(2):375-393. doi: 10.1111/pbi.13473. Epub 2020 Sep 20. Plant Biotechnol J. 2021. PMID: 32888338 Free PMC article.
References
-
- Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 - PMC - PubMed
-
- Arimura G.I., Ozawa R., Kugimiya S., Takabayashi J., Bohlmann J. (2004). Herbivore-induced defense response in a model legume. Two-spotted spider mites induce emission of (E)-beta-ocimene and transcript accumulation of (E)-beta-ocimene synthase in Lotus japonicus. Plant Physiol. 135: 1976–1983 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

