FastML: a web server for probabilistic reconstruction of ancestral sequences
- PMID: 22661579
- PMCID: PMC3394241
- DOI: 10.1093/nar/gks498
FastML: a web server for probabilistic reconstruction of ancestral sequences
Abstract
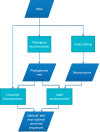
Ancestral sequence reconstruction is essential to a variety of evolutionary studies. Here, we present the FastML web server, a user-friendly tool for the reconstruction of ancestral sequences. FastML implements various novel features that differentiate it from existing tools: (i) FastML uses an indel-coding method, in which each gap, possibly spanning multiples sites, is coded as binary data. FastML then reconstructs ancestral indel states assuming a continuous time Markov process. FastML provides the most likely ancestral sequences, integrating both indels and characters; (ii) FastML accounts for uncertainty in ancestral states: it provides not only the posterior probabilities for each character and indel at each sequence position, but also a sample of ancestral sequences from this posterior distribution, and a list of the k-most likely ancestral sequences; (iii) FastML implements a large array of evolutionary models, which makes it generic and applicable for nucleotide, protein and codon sequences; and (iv) a graphical representation of the results is provided, including, for example, a graphical logo of the inferred ancestral sequences. The utility of FastML is demonstrated by reconstructing ancestral sequences of the Env protein from various HIV-1 subtypes. FastML is freely available for all academic users and is available online at http://fastml.tau.ac.il/.
Figures

Similar articles
-
ARPIP: Ancestral Sequence Reconstruction with Insertions and Deletions under the Poisson Indel Process.Syst Biol. 2023 Jun 16;72(2):307-318. doi: 10.1093/sysbio/syac050. Syst Biol. 2023. PMID: 35866991 Free PMC article.
-
GUIDANCE: a web server for assessing alignment confidence scores.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W23-8. doi: 10.1093/nar/gkq443. Epub 2010 May 23. Nucleic Acids Res. 2010. PMID: 20497997 Free PMC article.
-
Indel reliability in indel-based phylogenetic inference.Genome Biol Evol. 2014 Nov 18;6(12):3199-209. doi: 10.1093/gbe/evu252. Genome Biol Evol. 2014. PMID: 25409663 Free PMC article.
-
Please Mind the Gap: Indel-Aware Parsimony for Fast and Accurate Ancestral Sequence Reconstruction and Multiple Sequence Alignment Including Long Indels.Mol Biol Evol. 2024 Jul 3;41(7):msae109. doi: 10.1093/molbev/msae109. Mol Biol Evol. 2024. PMID: 38842253 Free PMC article.
-
webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser.BMC Bioinformatics. 2010 Nov 26;11:579. doi: 10.1186/1471-2105-11-579. BMC Bioinformatics. 2010. PMID: 21110866 Free PMC article.
Cited by
-
Novel insights into the origin and diversification of photosynthesis based on analyses of conserved indels in the core reaction center proteins.Photosynth Res. 2017 Feb;131(2):159-171. doi: 10.1007/s11120-016-0307-1. Epub 2016 Sep 16. Photosynth Res. 2017. PMID: 27638319
-
Statistical framework to determine indel-length distribution.Bioinformatics. 2024 Feb 1;40(2):btae043. doi: 10.1093/bioinformatics/btae043. Bioinformatics. 2024. PMID: 38269647 Free PMC article.
-
Alignment-Integrated Reconstruction of Ancestral Sequences Improves Accuracy.Genome Biol Evol. 2020 Sep 1;12(9):1549-1565. doi: 10.1093/gbe/evaa164. Genome Biol Evol. 2020. PMID: 32785673 Free PMC article.
-
Ancestral sequence reconstruction - An underused approach to understand the evolution of gene function in plants?Comput Struct Biotechnol J. 2021 Mar 16;19:1579-1594. doi: 10.1016/j.csbj.2021.03.008. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33868595 Free PMC article. Review.
-
Recurrent evolution of an inhibitor of ESCRT-dependent virus budding and LINE-1 retrotransposition in primates.Curr Biol. 2022 Apr 11;32(7):1511-1522.e6. doi: 10.1016/j.cub.2022.02.018. Epub 2022 Mar 3. Curr Biol. 2022. PMID: 35245459 Free PMC article.
References
-
- Liberles DA. Ancestral Sequence Reconstruction. Oxford: Oxford University Press; 2007.
-
- Chang BS, Jonsson K, Kazmi MA, Donoghue MJ, Sakmar TP. Recreating a functional ancestral archosaur visual pigment. Mol. Biol. Evol. 2002;19:1483–1489. - PubMed
-
- Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–1717. - PubMed
-
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

