Novel therapeutic targets in the brain tumor microenvironment
- PMID: 22643827
- PMCID: PMC3388186
- DOI: 10.18632/oncotarget.526
Novel therapeutic targets in the brain tumor microenvironment
Abstract
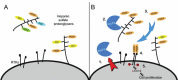
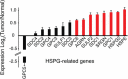
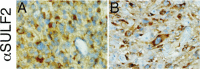
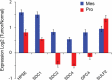
Glioblastoma (GBM), a highly malignant brain tumor of adults and children, diffusely invades within the non-neoplastic brain. Despite aggressive current therapeutic interventions, improved therapeutic strategies are greatly needed. Interactions between the tumor and constituents of its microenvironment are known to regulate malignancy, and heparan sulfate proteoglycans (HSPGs) are important as they bind diverse extracellular proteins, including growth factors and cell adhesion molecules, regulating the activity of several ligand-mediated signaling pathways. Recent work from our group described a mechanism by which GBM regulates PDGFR-alpha signaling via enzymatic alteration of heparan sulfate proteoglycans (HSPGs) in the extracellular microenvironment. Blocking tumor-induced alterations of HSPGs, which can be achieved by pharmacological strategies, would potentially inhibit multiple oncogenic signaling pathways in tumor cells and disrupt critical tumormicroenvironment interactions. Here we examine HSPGs and the enzymes that modify them in GBM. We compare their expression across tumor subtypes, their potential roles in oncogenesis, and their potential as novel therapeutic targets in GBM.
Figures
Similar articles
-
Epigenetic Regulation of the Biosynthesis & Enzymatic Modification of Heparan Sulfate Proteoglycans: Implications for Tumorigenesis and Cancer Biomarkers.Int J Mol Sci. 2017 Jun 26;18(7):1361. doi: 10.3390/ijms18071361. Int J Mol Sci. 2017. PMID: 28672878 Free PMC article. Review.
-
Heparan Sulfate Proteoglycan Signaling in Tumor Microenvironment.Int J Mol Sci. 2020 Sep 9;21(18):6588. doi: 10.3390/ijms21186588. Int J Mol Sci. 2020. PMID: 32916872 Free PMC article. Review.
-
Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: growth regulation and the prospect of new cancer therapies.J Cell Biochem. 2005 Dec 1;96(5):897-905. doi: 10.1002/jcb.20602. J Cell Biochem. 2005. PMID: 16149080 Review.
-
Heparan Sulfate Glycosaminoglycans in Glioblastoma Promote Tumor Invasion.Mol Cancer Res. 2017 Nov;15(11):1623-1633. doi: 10.1158/1541-7786.MCR-17-0352. Epub 2017 Aug 4. Mol Cancer Res. 2017. PMID: 28778876 Free PMC article.
-
Heparan Sulfate in the Tumor Microenvironment.Adv Exp Med Biol. 2020;1245:147-161. doi: 10.1007/978-3-030-40146-7_7. Adv Exp Med Biol. 2020. PMID: 32266657 Review.
Cited by
-
Proteoglycans and their roles in brain cancer.FEBS J. 2013 May;280(10):2399-417. doi: 10.1111/febs.12109. Epub 2013 Feb 6. FEBS J. 2013. PMID: 23281850 Free PMC article. Review.
-
Chemistry and Function of Glycosaminoglycans in the Nervous System.Adv Neurobiol. 2023;29:117-162. doi: 10.1007/978-3-031-12390-0_5. Adv Neurobiol. 2023. PMID: 36255674 Review.
-
Targeting glioblastoma with NK cells and mAb against NG2/CSPG4 prolongs animal survival.Oncotarget. 2013 Sep;4(9):1527-46. doi: 10.18632/oncotarget.1291. Oncotarget. 2013. PMID: 24127551 Free PMC article.
-
In-Depth Matrisome and Glycoproteomic Analysis of Human Brain Glioblastoma Versus Control Tissue.Mol Cell Proteomics. 2022 Apr;21(4):100216. doi: 10.1016/j.mcpro.2022.100216. Epub 2022 Feb 23. Mol Cell Proteomics. 2022. PMID: 35202840 Free PMC article.
-
Serglycin activates pro-tumorigenic signaling and controls glioblastoma cell stemness, differentiation and invasive potential.Matrix Biol Plus. 2020 Mar 11;6-7:100033. doi: 10.1016/j.mbplus.2020.100033. eCollection 2020 May. Matrix Biol Plus. 2020. PMID: 33543029 Free PMC article.
References
-
- Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, Cloughesy TF, Nelson SF. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22(15):2361–2373. - PubMed
-
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. - PubMed
-
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

