Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1
- PMID: 22623528
- PMCID: PMC3386053
- DOI: 10.1073/pnas.1202924109
Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1
Abstract
Hepatitis C virus (HCV) infects more than 2% of the global population and is a leading cause of liver cirrhosis, hepatocellular carcinoma, and end-stage liver diseases. Circulating HCV is genetically diverse, and therefore a broadly effective vaccine must target conserved T- and B-cell epitopes of the virus. Human mAb HCV1 has broad neutralizing activity against HCV isolates from at least four major genotypes and protects in the chimpanzee model from primary HCV challenge. The antibody targets a conserved antigenic site (residues 412-423) on the virus E2 envelope glycoprotein. Two crystal structures of HCV1 Fab in complex with an epitope peptide at 1.8-Å resolution reveal that the epitope is a β-hairpin displaying a hydrophilic face and a hydrophobic face on opposing sides of the hairpin. The antibody predominantly interacts with E2 residues Leu(413) and Trp(420) on the hydrophobic face of the epitope, thus providing an explanation for how HCV isolates bearing mutations at Asn(415) on the same binding face escape neutralization by this antibody. The results provide structural information for a neutralizing epitope on the HCV E2 glycoprotein and should help guide rational design of HCV immunogens to elicit similar broadly neutralizing antibodies through vaccination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


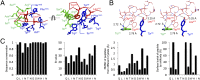
Comment in
-
Unraveling hepatitis C virus structure.Cell Res. 2014 Apr;24(4):385-6. doi: 10.1038/cr.2014.31. Epub 2014 Mar 14. Cell Res. 2014. PMID: 24626133 Free PMC article.
Similar articles
-
Structural basis for penetration of the glycan shield of hepatitis C virus E2 glycoprotein by a broadly neutralizing human antibody.J Biol Chem. 2015 Apr 17;290(16):10117-25. doi: 10.1074/jbc.M115.643528. Epub 2015 Mar 3. J Biol Chem. 2015. PMID: 25737449 Free PMC article.
-
Structure-Based Design of Hepatitis C Virus Vaccines That Elicit Neutralizing Antibody Responses to a Conserved Epitope.J Virol. 2017 Sep 27;91(20):e01032-17. doi: 10.1128/JVI.01032-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28794021 Free PMC article.
-
Escape of Hepatitis C Virus from Epitope I Neutralization Increases Sensitivity of Other Neutralization Epitopes.J Virol. 2018 Apr 13;92(9):e02066-17. doi: 10.1128/JVI.02066-17. Print 2018 May 1. J Virol. 2018. PMID: 29467319 Free PMC article.
-
HCV Glycoprotein Structure and Implications for B-Cell Vaccine Development.Int J Mol Sci. 2020 Sep 16;21(18):6781. doi: 10.3390/ijms21186781. Int J Mol Sci. 2020. PMID: 32947858 Free PMC article. Review.
-
Mapping Determinants of Virus Neutralization and Viral Escape for Rational Design of a Hepatitis C Virus Vaccine.Front Immunol. 2018 May 31;9:1194. doi: 10.3389/fimmu.2018.01194. eCollection 2018. Front Immunol. 2018. PMID: 29904384 Free PMC article. Review.
Cited by
-
Viral evasion and challenges of hepatitis C virus vaccine development.Curr Opin Virol. 2016 Oct;20:55-63. doi: 10.1016/j.coviro.2016.09.004. Epub 2016 Sep 19. Curr Opin Virol. 2016. PMID: 27657659 Free PMC article. Review.
-
The secret life of viral entry glycoproteins: moonlighting in immune evasion.PLoS Pathog. 2013;9(5):e1003258. doi: 10.1371/journal.ppat.1003258. Epub 2013 May 16. PLoS Pathog. 2013. PMID: 23696729 Free PMC article. Review. No abstract available.
-
HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.Cell Host Microbe. 2018 Nov 14;24(5):703-716.e3. doi: 10.1016/j.chom.2018.10.009. Cell Host Microbe. 2018. PMID: 30439340 Free PMC article.
-
A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes.Immunology. 2014 Sep;143(1):13-20. doi: 10.1111/imm.12323. Immunology. 2014. PMID: 24854488 Free PMC article. Review.
-
Non-random escape pathways from a broadly neutralizing human monoclonal antibody map to a highly conserved region on the hepatitis C virus E2 glycoprotein encompassing amino acids 412-423.PLoS Pathog. 2014 Aug 14;10(8):e1004297. doi: 10.1371/journal.ppat.1004297. eCollection 2014 Aug. PLoS Pathog. 2014. PMID: 25122476 Free PMC article.
References
-
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. - PubMed
-
- Wasley A, Miller JT, Finelli L. Centers for Disease Control and Prevention (CDC) Surveillance for acute viral hepatitis—United States, 2005. MMWR Surveill Summ. 2007;56:1–24. - PubMed
-
- Onofrey S, et al. Centers for Disease Control and Prevention (CDC) Hepatitis C virus infection among adolescents and young adults: Massachusetts, 2002-2009. MMWR Morb Mortal Wkly Rep. 2011;60:537–541. - PubMed
-
- Lemon SM, Walker C, Alter MJ, Yi M. In: Hepatitis C Virus. Virology. 5th Ed. Knipe DM, et al., editors. Vol 1. Philadelphia: Lippincott-Raven; 2007. pp. 1253–1304.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

