Specialized ribosomes: a new frontier in gene regulation and organismal biology
- PMID: 22617470
- PMCID: PMC4039366
- DOI: 10.1038/nrm3359
Specialized ribosomes: a new frontier in gene regulation and organismal biology
Abstract
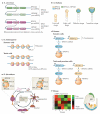
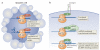
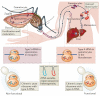
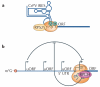
Historically, the ribosome has been viewed as a complex ribozyme with constitutive rather than intrinsic regulatory capacity in mRNA translation. However, emerging studies reveal that ribosome activity may be highly regulated. Heterogeneity in ribosome composition resulting from differential expression and post-translational modifications of ribosomal proteins, ribosomal RNA (rRNA) diversity and the activity of ribosome-associated factors may generate 'specialized ribosomes' that have a substantial impact on how the genomic template is translated into functional proteins. Moreover, constitutive components of the ribosome may also exert more specialized activities by virtue of their interactions with specific mRNA regulatory elements such as internal ribosome entry sites (IRESs) or upstream open reading frames (uORFs). Here we discuss the hypothesis that intrinsic regulation by the ribosome acts to selectively translate subsets of mRNAs harbouring unique cis-regulatory elements, thereby introducing an additional level of regulation in gene expression and the life of an organism.
Figures

Similar articles
-
The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life.Mol Cell. 2018 Aug 2;71(3):364-374. doi: 10.1016/j.molcel.2018.07.018. Mol Cell. 2018. PMID: 30075139 Free PMC article. Review.
-
Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide.Mol Cell. 2017 Jul 6;67(1):71-83.e7. doi: 10.1016/j.molcel.2017.05.021. Epub 2017 Jun 15. Mol Cell. 2017. PMID: 28625553 Free PMC article.
-
Translation regulation by ribosomes: Increased complexity and expanded scope.RNA Biol. 2016 Sep;13(9):748-55. doi: 10.1080/15476286.2015.1107701. Epub 2015 Oct 29. RNA Biol. 2016. PMID: 26513496 Free PMC article. Review.
-
Emerging Role of Eukaryote Ribosomes in Translational Control.Int J Mol Sci. 2019 Mar 11;20(5):1226. doi: 10.3390/ijms20051226. Int J Mol Sci. 2019. PMID: 30862090 Free PMC article. Review.
-
Cis-regulatory RNA elements that regulate specialized ribosome activity.RNA Biol. 2015;12(10):1083-7. doi: 10.1080/15476286.2015.1085149. Epub 2015 Sep 1. RNA Biol. 2015. PMID: 26327194 Free PMC article. Review.
Cited by
-
Translational control in cancer etiology.Cold Spring Harb Perspect Biol. 2013 Feb 1;5(2):a012336. doi: 10.1101/cshperspect.a012336. Cold Spring Harb Perspect Biol. 2013. PMID: 22767671 Free PMC article. Review.
-
Low dose ribosomal DNA P-loop mutation affects development and enforces autophagy in Arabidopsis.RNA Biol. 2024 Jan;21(1):1-15. doi: 10.1080/15476286.2023.2298532. Epub 2023 Dec 29. RNA Biol. 2024. PMID: 38156797 Free PMC article.
-
Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors.Proc Natl Acad Sci U S A. 2012 Nov 27;109(48):19537-44. doi: 10.1073/pnas.1214774109. Epub 2012 Nov 9. Proc Natl Acad Sci U S A. 2012. PMID: 23144218 Free PMC article.
-
Human PDCD2L Is an Export Substrate of CRM1 That Associates with 40S Ribosomal Subunit Precursors.Mol Cell Biol. 2016 Nov 28;36(24):3019-3032. doi: 10.1128/MCB.00303-16. Print 2016 Dec 15. Mol Cell Biol. 2016. PMID: 27697862 Free PMC article.
-
Evidence of non-tandemly repeated rDNAs and their intragenomic heterogeneity in Rhizophagus irregularis.Commun Biol. 2018 Jul 10;1:87. doi: 10.1038/s42003-018-0094-7. eCollection 2018. Commun Biol. 2018. PMID: 30271968 Free PMC article.
References
-
- Frank J. The ribosome — a macromolecular machine par excellence. Chem. Biol. 2000;7:R133–R141. - PubMed
-
-
Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. Reviews the mechanism of translation initiation and its regulation..
-
-
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. - PubMed
-
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

