Structural basis for the acyltransferase activity of lecithin:retinol acyltransferase-like proteins
- PMID: 22605381
- PMCID: PMC3390653
- DOI: 10.1074/jbc.M112.361550
Structural basis for the acyltransferase activity of lecithin:retinol acyltransferase-like proteins
Abstract
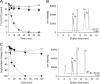
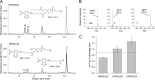
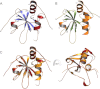
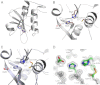
Lecithin:retinol acyltransferase-like proteins, also referred to as HRAS-like tumor suppressors, comprise a vertebrate subfamily of papain-like or NlpC/P60 thiol proteases that function as phospholipid-metabolizing enzymes. HRAS-like tumor suppressor 3, a representative member of this group, plays a key role in regulating triglyceride accumulation and energy expenditure in adipocytes and therefore constitutes a novel pharmacological target for treatment of metabolic disorders causing obesity. Here, we delineate a catalytic mechanism common to lecithin:retinol acyltransferase-like proteins and provide evidence for their alternative robust lipid-dependent acyltransferase enzymatic activity. We also determined high resolution crystal structures of HRAS-like tumor suppressor 2 and 3 to gain insight into their active site architecture. Based on this structural analysis, two conformational states of the catalytic Cys-113 were identified that differ in reactivity and thus could define the catalytic properties of these two proteins. Finally, these structures provide a model for the topology of these enzymes and allow identification of the protein-lipid bilayer interface. This study contributes to the enzymatic and structural understanding of HRAS-like tumor suppressor enzymes.
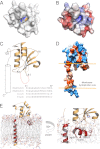
Figures




Similar articles
-
Structure/function relationships of adipose phospholipase A2 containing a cys-his-his catalytic triad.J Biol Chem. 2012 Oct 12;287(42):35260-35274. doi: 10.1074/jbc.M112.398859. Epub 2012 Aug 25. J Biol Chem. 2012. PMID: 22923616 Free PMC article.
-
LRAT-specific domain facilitates vitamin A metabolism by domain swapping in HRASLS3.Nat Chem Biol. 2015 Jan;11(1):26-32. doi: 10.1038/nchembio.1687. Epub 2014 Nov 10. Nat Chem Biol. 2015. PMID: 25383759 Free PMC article.
-
Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes.Biochim Biophys Acta. 2009 Dec;1791(12):1114-24. doi: 10.1016/j.bbalip.2009.07.001. Epub 2009 Jul 14. Biochim Biophys Acta. 2009. PMID: 19615464
-
The HRASLS (PLA/AT) subfamily of enzymes.J Biomed Sci. 2015 Oct 26;22:99. doi: 10.1186/s12929-015-0210-7. J Biomed Sci. 2015. PMID: 26503625 Free PMC article. Review.
-
Membrane-bound lecithin-retinol acyltransferase.Biochem Biophys Res Commun. 2002 Apr 19;292(5):1243-50. doi: 10.1006/bbrc.2001.2021. Biochem Biophys Res Commun. 2002. PMID: 11969222 Review. No abstract available.
Cited by
-
Membrane enzymes: transformers at the interface.Nat Chem Biol. 2015 Feb;11(2):102-3. doi: 10.1038/nchembio.1738. Epub 2015 Jan 12. Nat Chem Biol. 2015. PMID: 25580855 Review. No abstract available.
-
Current Knowledge on Mammalian Phospholipase A1, Brief History, Structures, Biochemical and Pathophysiological Roles.Molecules. 2022 Apr 12;27(8):2487. doi: 10.3390/molecules27082487. Molecules. 2022. PMID: 35458682 Free PMC article. Review.
-
PLA2G16 represents a switch between entry and clearance of Picornaviridae.Nature. 2017 Jan 19;541(7637):412-416. doi: 10.1038/nature21032. Epub 2017 Jan 11. Nature. 2017. PMID: 28077878
-
Chemistry and biology of the initial steps in vision: the Friedenwald lecture.Invest Ophthalmol Vis Sci. 2014 Oct 22;55(10):6651-72. doi: 10.1167/iovs.14-15502. Invest Ophthalmol Vis Sci. 2014. PMID: 25338686 Free PMC article.
-
Endocannabinoids, related compounds and their metabolic routes.Molecules. 2014 Oct 24;19(11):17078-106. doi: 10.3390/molecules191117078. Molecules. 2014. PMID: 25347455 Free PMC article. Review.
References
-
- Fernandis A. Z., Wenk M. R. (2007) Membrane lipids as signaling molecules. Curr. Opin. Lipidol. 18, 121–128 - PubMed
-
- Hannun Y. A., Obeid L. M. (2008) Principles of bioactive lipid signaling. Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150 - PubMed
-
- Ruiz A., Winston A., Lim Y. H., Gilbert B. A., Rando R. R., Bok D. (1999) Molecular and biochemical characterization of lecithin-retinol acyltransferase. J. Biol. Chem. 274, 3834–3841 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- P41 RR012408/RR/NCRR NIH HHS/United States
- R01 EY009339/EY/NEI NIH HHS/United States
- 8 P41 GM103403-10/GM/NIGMS NIH HHS/United States
- P41 RR015301/RR/NCRR NIH HHS/United States
- P41 GM103473/GM/NIGMS NIH HHS/United States
- DK079221/DK/NIDDK NIH HHS/United States
- R01 EY008061/EY/NEI NIH HHS/United States
- R00 EY019718/EY/NEI NIH HHS/United States
- EY019718/EY/NEI NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- 5P41RR015301-10/RR/NCRR NIH HHS/United States
- EY009339/EY/NEI NIH HHS/United States
- EY008061/EY/NEI NIH HHS/United States
- K99 EY019718/EY/NEI NIH HHS/United States
- R01 DK079221/DK/NIDDK NIH HHS/United States
- P41 GM103403/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

