Somatic STAT3 mutations in large granular lymphocytic leukemia
- PMID: 22591296
- PMCID: PMC3693860
- DOI: 10.1056/NEJMoa1114885
Somatic STAT3 mutations in large granular lymphocytic leukemia
Abstract
Background: T-cell large granular lymphocytic leukemia is a rare lymphoproliferative disorder characterized by the expansion of clonal CD3+CD8+ cytotoxic T lymphocytes (CTLs) and often associated with autoimmune disorders and immune-mediated cytopenias.
Methods: We used next-generation exome sequencing to identify somatic mutations in CTLs from an index patient with large granular lymphocytic leukemia. Targeted resequencing was performed in a well-characterized cohort of 76 patients with this disorder, characterized by clonal T-cell-receptor rearrangements and increased numbers of large granular lymphocytes.
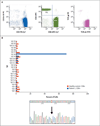
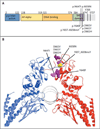
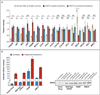
Results: Mutations in the signal transducer and activator of transcription 3 gene (STAT3) were found in 31 of 77 patients (40%) with large granular lymphocytic leukemia. Among these 31 patients, recurrent mutational hot spots included Y640F in 13 (17%), D661V in 7 (9%), D661Y in 7 (9%), and N647I in 3 (4%). All mutations were located in exon 21, encoding the Src homology 2 (SH2) domain, which mediates the dimerization and activation of STAT protein. The amino acid changes resulted in a more hydrophobic protein surface and were associated with phosphorylation of STAT3 and its localization in the nucleus. In vitro functional studies showed that the Y640F and D661V mutations increased the transcriptional activity of STAT3. In the affected patients, downstream target genes of the STAT3 pathway (IFNGR2, BCL2L1, and JAK2) were up-regulated. Patients with STAT3 mutations presented more often with neutropenia and rheumatoid arthritis than did patients without these mutations.
Conclusions: The SH2 dimerization and activation domain of STAT3 is frequently mutated in patients with large granular lymphocytic leukemia; these findings suggest that aberrant STAT3 signaling underlies the pathogenesis of this disease. (Funded by the Academy of Finland and others.).
Figures

Similar articles
-
Uncovering the pathogenesis of large granular lymphocytic leukemia-novel STAT3 and STAT5b mutations.Ann Med. 2014 May;46(3):114-22. doi: 10.3109/07853890.2014.882105. Epub 2014 Feb 11. Ann Med. 2014. PMID: 24512550 Review.
-
STAT3 gene mutations and their association with pure red cell aplasia in large granular lymphocyte leukemia.Cancer Sci. 2014 Mar;105(3):342-6. doi: 10.1111/cas.12341. Epub 2014 Jan 22. Cancer Sci. 2014. PMID: 24350896 Free PMC article.
-
Somatic STAT3 mutations in Felty syndrome: an implication for a common pathogenesis with large granular lymphocyte leukemia.Haematologica. 2018 Feb;103(2):304-312. doi: 10.3324/haematol.2017.175729. Epub 2017 Dec 7. Haematologica. 2018. PMID: 29217783 Free PMC article.
-
The analysis of clonal diversity and therapy responses using STAT3 mutations as a molecular marker in large granular lymphocytic leukemia.Haematologica. 2015 Jan;100(1):91-9. doi: 10.3324/haematol.2014.113142. Epub 2014 Oct 3. Haematologica. 2015. PMID: 25281507 Free PMC article.
-
[Large Granular Lymphocytic Leukemia and JAK/STAT Signaling Pathway--Review].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2016 Feb;24(1):254-60. doi: 10.7534/j.issn.1009-2137.2016.01.049. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2016. PMID: 26913432 Review. Chinese.
Cited by
-
STAT3 mutations identified in human hematologic neoplasms induce myeloid malignancies in a mouse bone marrow transplantation model.Haematologica. 2013 Nov;98(11):1748-52. doi: 10.3324/haematol.2013.085068. Epub 2013 Jul 19. Haematologica. 2013. PMID: 23872306 Free PMC article.
-
STAT3 mutations in "gray-zone" cases of T-cell large granular lymphocytic leukemia associated with autoimmune rheumatic diseases.Front Med (Lausanne). 2022 Aug 31;9:1000265. doi: 10.3389/fmed.2022.1000265. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36117975 Free PMC article.
-
The constitutive activation of STAT3 gene and its mutations are at the crossroad between LGL leukemia and autoimmune disorders.Blood Cancer J. 2024 Jan 18;14(1):13. doi: 10.1038/s41408-024-00977-0. Blood Cancer J. 2024. PMID: 38238319 Free PMC article. Review.
-
Molecular Features and Diagnostic Challenges in Alpha/Beta T-Cell Large Granular Lymphocyte Leukemia.Int J Mol Sci. 2022 Nov 2;23(21):13392. doi: 10.3390/ijms232113392. Int J Mol Sci. 2022. PMID: 36362180 Free PMC article.
-
Pathophysiologic Mechanisms And Management Of Large Granular Lymphocytic Leukemia Associated Pure Red Cell Aplasia.Onco Targets Ther. 2019 Oct 4;12:8229-8240. doi: 10.2147/OTT.S222378. eCollection 2019. Onco Targets Ther. 2019. PMID: 31632073 Free PMC article.
References
-
- Loughran TP, Jr., Kadin ME, Starkebaum G, et al. Leukemia of large granular lymphocytes: association with clonal chromosomal abnormalities and autoimmune neutropenia, thrombocytopenia, and hemolytic anemia. Ann Intern Med. 1985;102:169–175. - PubMed
-
- Loughran TP., Jr. Clonal diseases of large granular lymphocytes. Blood. 1993;82:1–14. - PubMed
-
- Sokol L, Loughran TP., Jr. Large granular lymphocyte leukemia. Oncologist. 2006;11:263–273. - PubMed
-
- Burks EJ, Loughran TP., Jr. Pathogenesis of neutropenia in large granular lymphocyte leukemia and Felty syndrome. Blood Rev. 2006;20:245–266. - PubMed
-
- Rossi D, Franceschetti S, Capello D, et al. Transient monoclonal expansion of CD8+/CD57+ T-cell large granular lymphocytes after primary cytomegalovirus infection. Am J Hematol. 2007;82:1103–1105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
